Introduction & Data Modalities
Welcome
- Syllabus
- There will be group projects
- Most of the things you learn will come from the projects
- Auditors – decide if you want to participate in the group projects.
Homework
- Map the Box folder for the course to your own Box account and setup Box to sync on your laptop
- Install FSL
Data repositories
- Reproducible Brain Charts – Primary dataset for this course
- Adolescent Brain Cognitive Development (ABCD)
Reproducible Brain Charts
- We will use structural, resting state functional MRI (rs-fMRI), and “task” fMRI (t-fMRI)
ABCD Study
- Adolescent Brain Cognitive Development (ABCD)
- Amazing resource for large-scale imaging
- Huge study, requires complex data use agreement
Software
Analysis software
- Statistical Parametric Mapping (SPM; in MATLAB)
- FSL (linux/terminal based) – Primary software for this course
- AFNI
- Neuroconductor – modular analysis packages in
R
Getting setup with FSL
- Visualize the FSL template image
- Visualize my brain
Visualization software
Visualization is as important in image analysis as univariate data analysis (Always look at your data)
- FSLeyes
- ITK-Snap
- Papaya
Rpackages,ggseg,ciftitools,fsbrain, see following reference- Chopra et al., 2023 – overview of reproducible tools
Brain anatomy and terminology
Anatomy and terminology
Directory paths will be relative to the course directory on Box.
- Gray Matter: Brain tissue composed mainly of neuronal cell bodies. It is involved in processing and interpreting information and is found in regions like the cortex and deep brain nuclei.
- White Matter: Tissue made up of axons coated with myelin, which appears white. White matter connects regions of gray matter, allowing communication across the brain.
- Ventricles: Through the middle of the brain filled with cerebrospinal fluid.
More brain anatomy and terminology
- Cortex: The outer layer of the brain’s gray matter. It is highly folded and responsible for higher cognitive functions like perception, decision-making, and voluntary movement.
- Subcortex: Brain regions located beneath the cortex, including structures like the thalamus, basal ganglia, and hippocampus. These areas are important for functions such as emotion, memory, and movement regulation.
- Cerebellum: A separate structure located at the back of the brain, underneath the cortex. It plays a key role in coordinating movement, balance, and fine motor skills.
Even more brain anatomy and terminology
- Lobes: The brain is divided into four main lobes — frontal, parietal, temporal, and occipital — each associated with specific functions.
- Gyri (singular: gyrus): Ridges on the brain’s surface.
- Sulci (singular: sulcus): Grooves between the gyri.
- Anterior / Posterior / Superior / Inferior: Anatomical directions — front/back/top/bottom.
Neuroimaging data
- Neuroimaging provide a way to study brain structure and function in vivo in humans
- Magnetic resonance imaging (MRI) is noninvasive
- “Possibly, the most promising technology to link human emotion, cognition, and behavior with the brain.” – me
What is an image?
- A function from I(v): \mathbb{R}^3 \mapsto \mathbb{R}.
- Numeric values in the image represent biological values.
- Visually brighter pixels are higher values.
- MRI are collected in the frequency domain, but few people analyze their data there
Medical imaging data formats
- DICOM: Medical image format, which includes a lot of metadata in the image header. Raw output from MRI scanner.
.dcm - NIfTI: File format for 3D or 4D neuroimaging data with less (but still a lot of) metadata in the image header.
.niior.nii.gz.
You don’t need to gunzip.nii.gz files, all software can open them directly.
Neuroimaging terminology
- Voxel: A 3D pixel.
- TR: Repetition time - time required to obtain an entire volume (typically for fMRI data).
- TE: Echo time - time between application of a radiofrequency (RF) pulse and the peak of the signal (echo) generated.
- EPI: Echo planar imaging - the type of imaging used to collect fMRI data.
- T1-weighted / T2-weighted images: Types of MRI scans for different tissue properties. T1 show anatomy with good contrast for gray and white matter; T2 highlight fluid and often used to detect pathology.
Orientation terminology
- Axial View: A horizontal slice of the brain, viewed from above (as if looking down from the top of the head).
- Coronal View: A vertical slice viewed from the front (as if looking straight at the face).
- Sagittal View: A vertical slice viewed from the side (as if looking at the brain in profile).
Image space
- MNI Space / Talairach Space: Standardized brain coordinate systems used to align and compare brain scans across individuals.
- Radiological versus neurological convention
- Radiological - (left on the right)
- Neurological - (right on the right)
- Nice overview on this stuff
- Radiological is more common
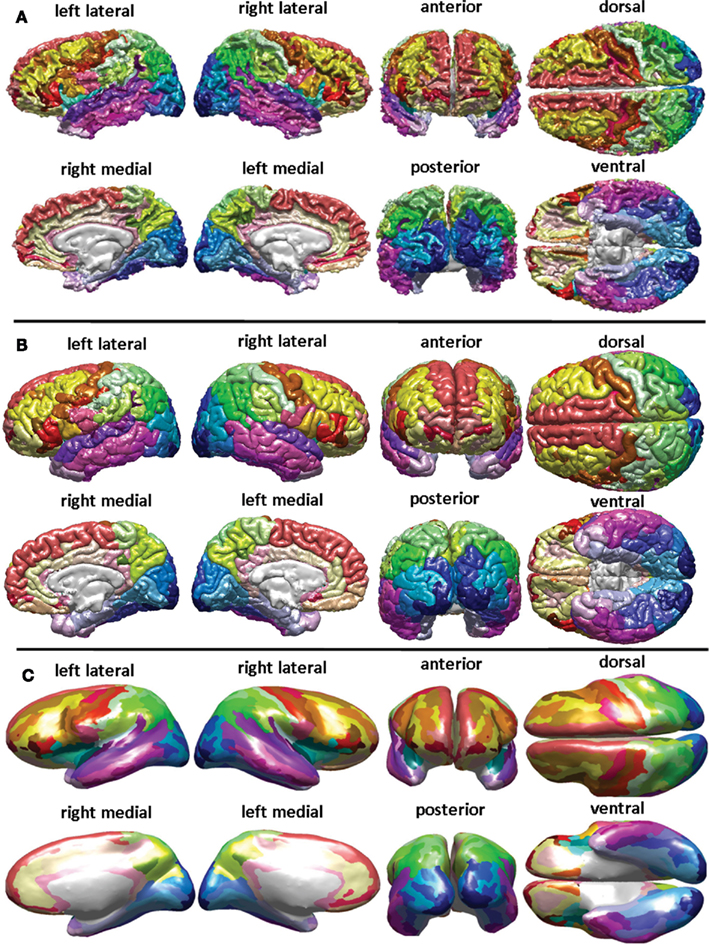
Atlases
Atlases define regions of the brain based on anatomical, microstructural, functional, or other features
- Common Atlases
- MNI
Data modalities
Structural imaging
Measures the anatomy of the brain
- Derived from T1/T2 MRI scans
- Structural measures
- Volume, Cortical thickness, Surface area
- Data structures
- Images
- Regions
- Gray matter surface
- Structural-based “similarity” networks
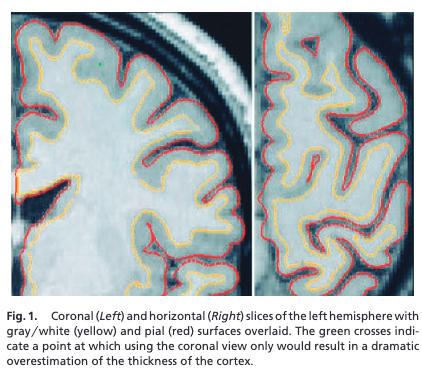
Freesurfer: structural analysis software
- Cortical surface estimation and analysis
- Now over 25 years old
- Many tools now built on analyzing data on the surface
Freesurfer visualization
Functional imaging
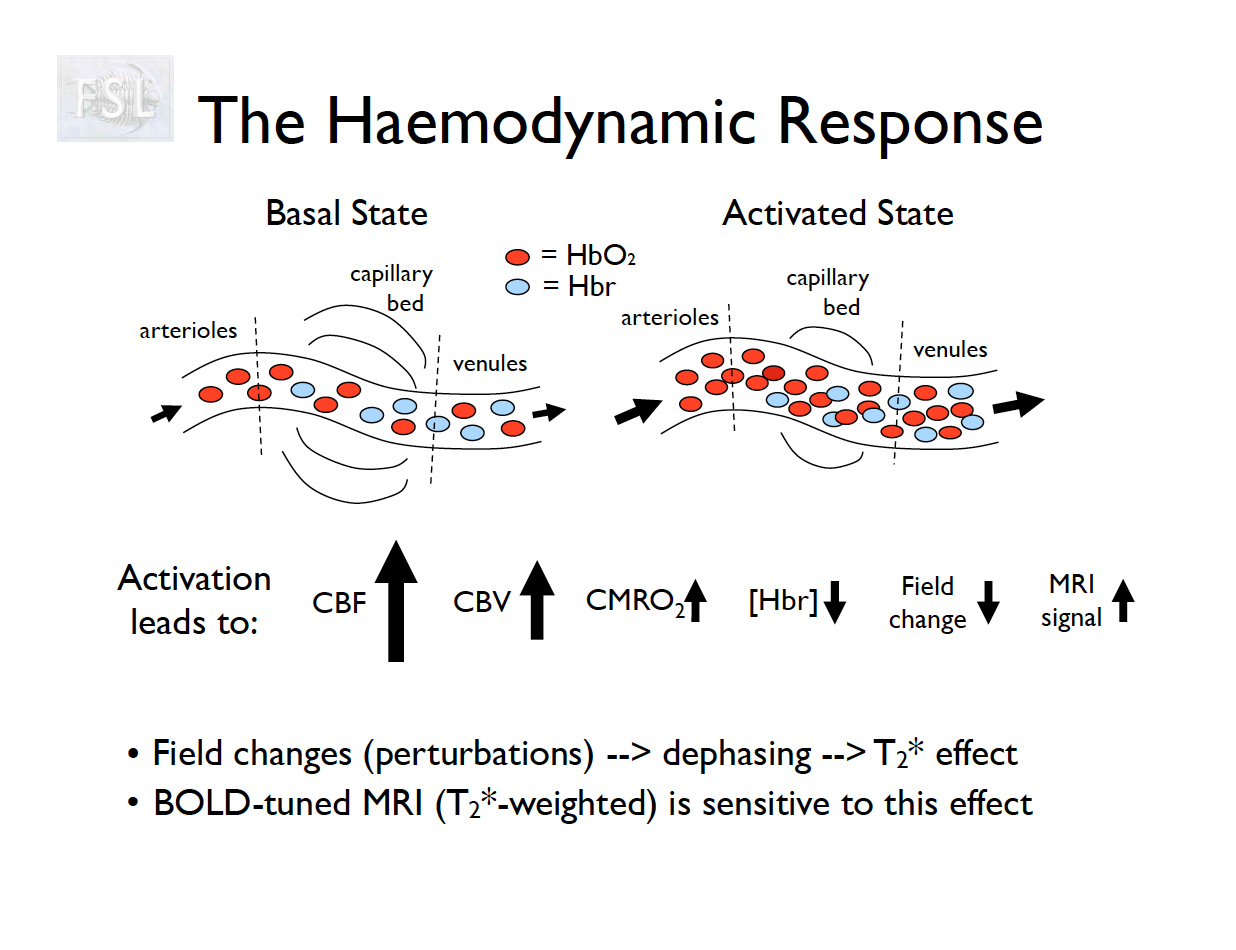
Measures brain functioning (inferred by deoxygentated blood)
- Blood oxygen level dependent (BOLD) activity
- History of functional MRI (Bandettini 2012)
- functional Magnetic Resonance Imaging (fMRI) are time-series data
Types of fMRI data
- task fMRI – acquired doing a task
- resting state fMRI (rs-fMRI) – acquired doing nothing
Task fMRI overview
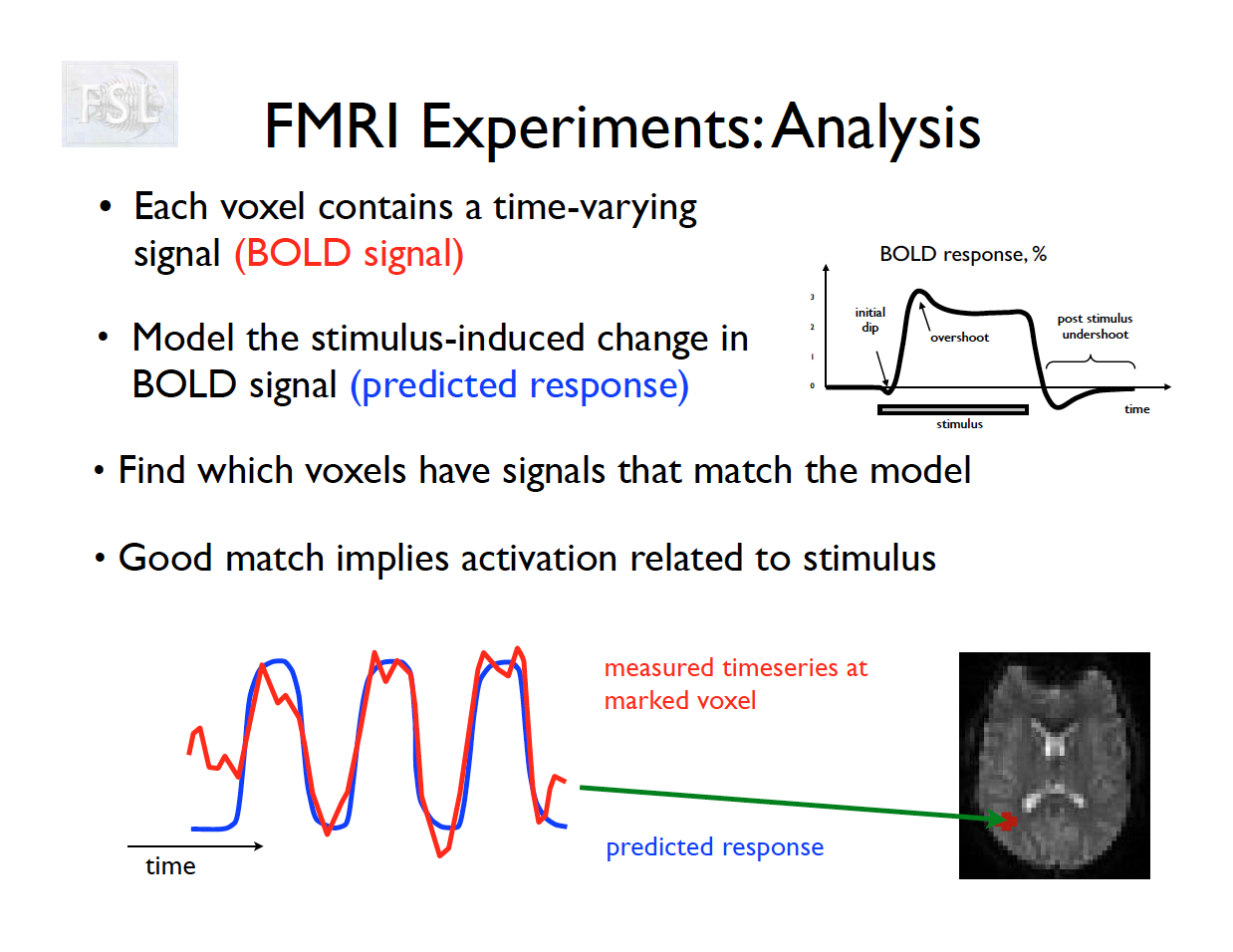
- Measures brain activity during a task using BOLD signal.
- Block design: repeated task periods alternating with baseline (e.g. 30s task, 30s rest).
- Event-related design: timing randomized, short stimuli.
- Naturalistic: E.g., viewing a movie.
- Analysis fits a time-series model and compares conditions.
- Data structures
- Images, Regions, Gray matter surface
Task fMRI design illustrations
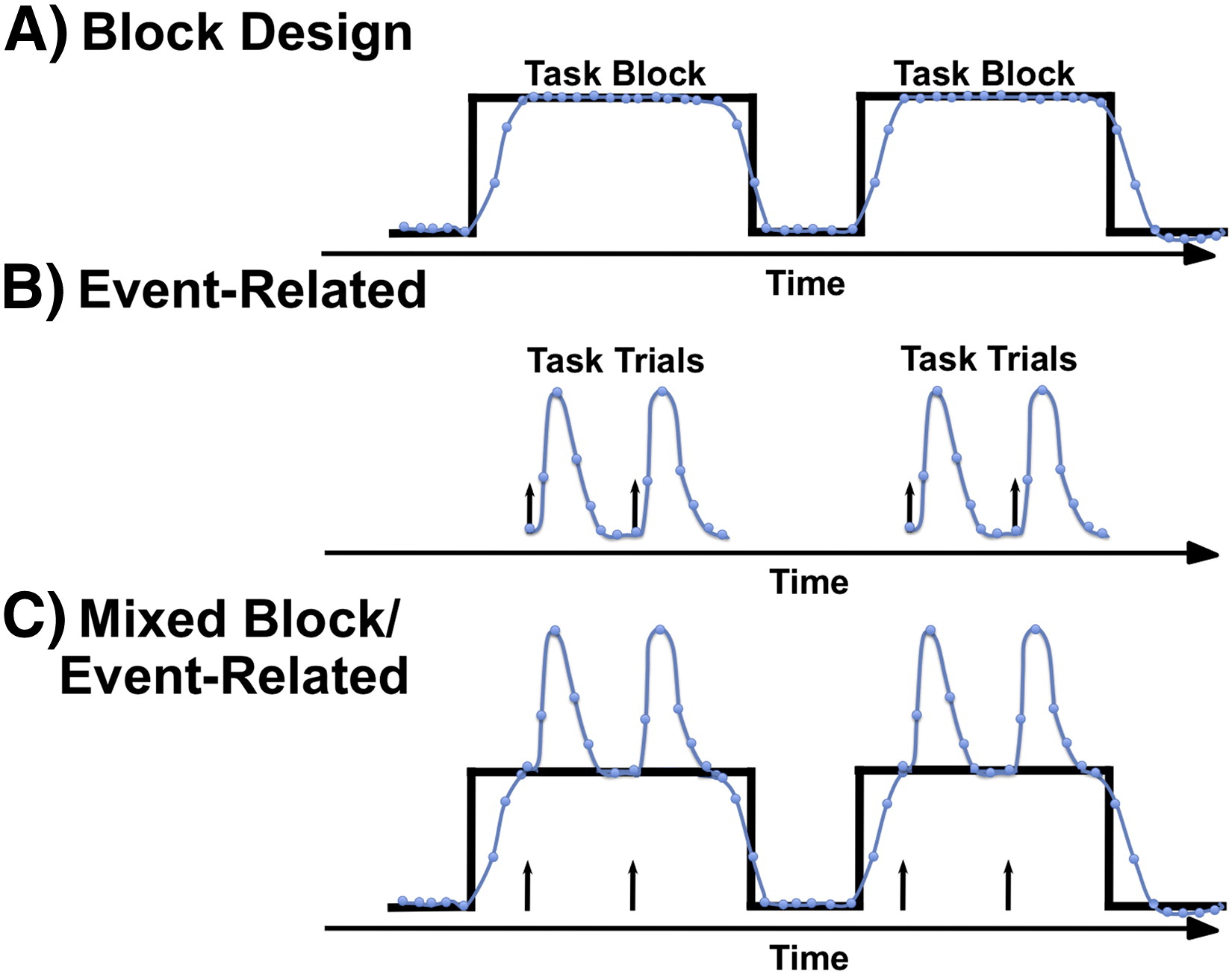
Block versus task designs Petersen & Dubis 2011
Notes on illustration
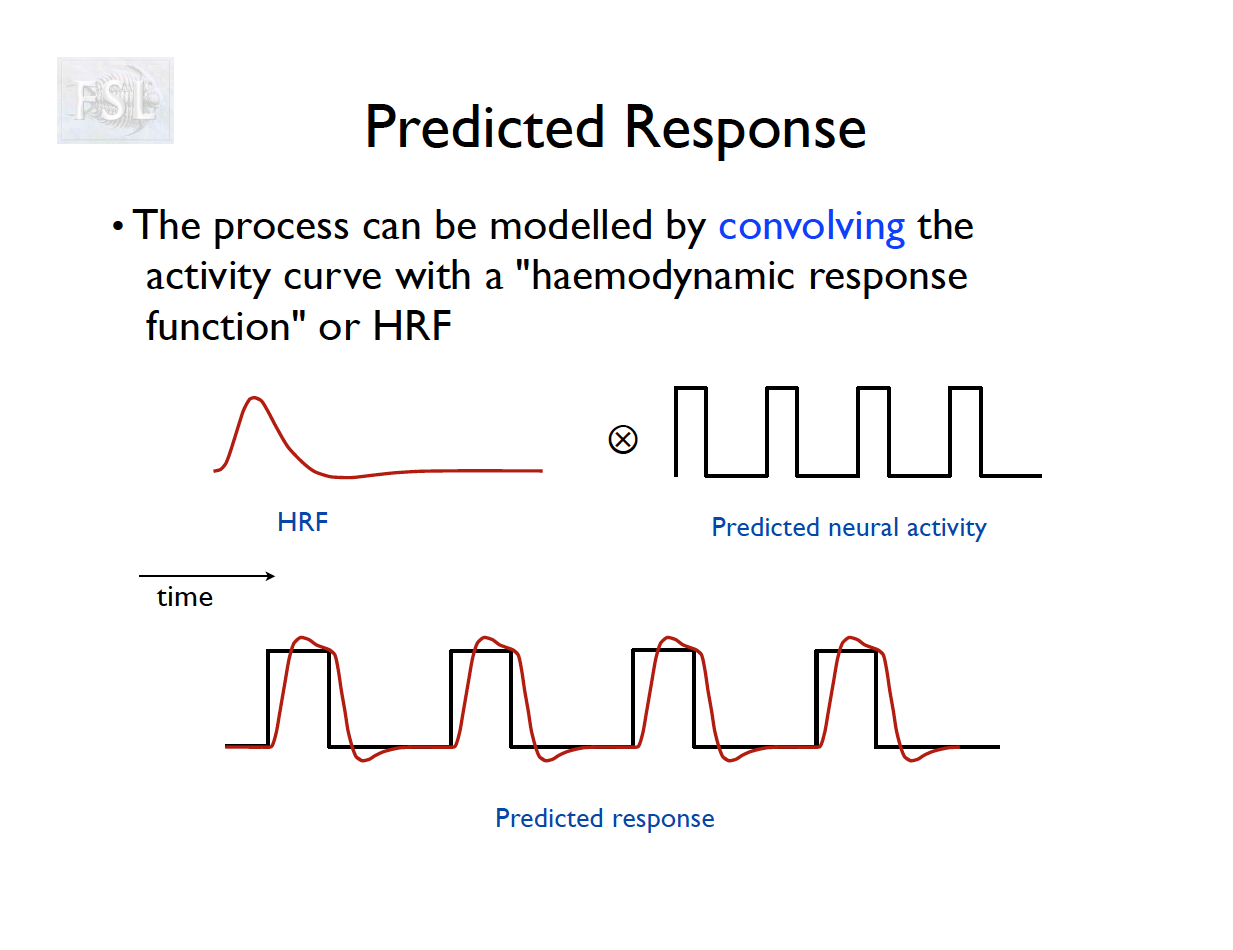
- Known stimulus time series are convolved with an assumed hemodynamic response function (HRF)
- HRF is well studied, often assumed, and likely inaccurate for many brain regions REFs
- Hemodynamic (blood) response to stimuli is slow.
- fMRI sampled every 1.5-3 seconds or so.
- RBC fMRI parameters
RBC task data
- We will analyze Naturalistic viewing data
- Naturalistic viewing of Despicable Me (Furtado 2024)
- Despicable Me clip on Netflix 1:02:09 - 1:12:09

rs-fMRI
- What it is? Correlation between fMRI time-series in the brain at rest (doing nothing).
- History: Biswal 1995 first study to do this
- Now, things are quite a bit more sophisticated
- Data structures
- Images
- Regions
- Gray matter surface
- Networks
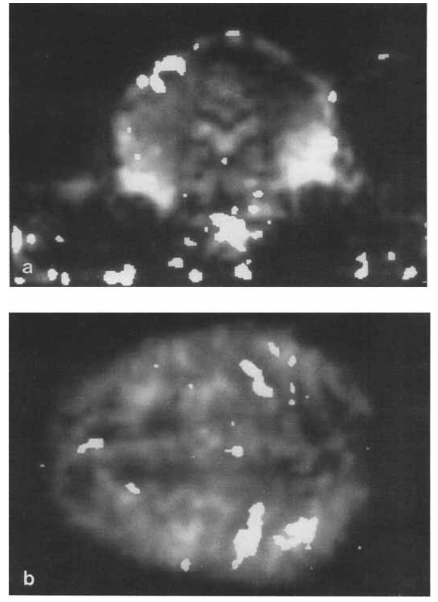
First resting state networks (De Luca)
Published in 2006 using independent components analysis
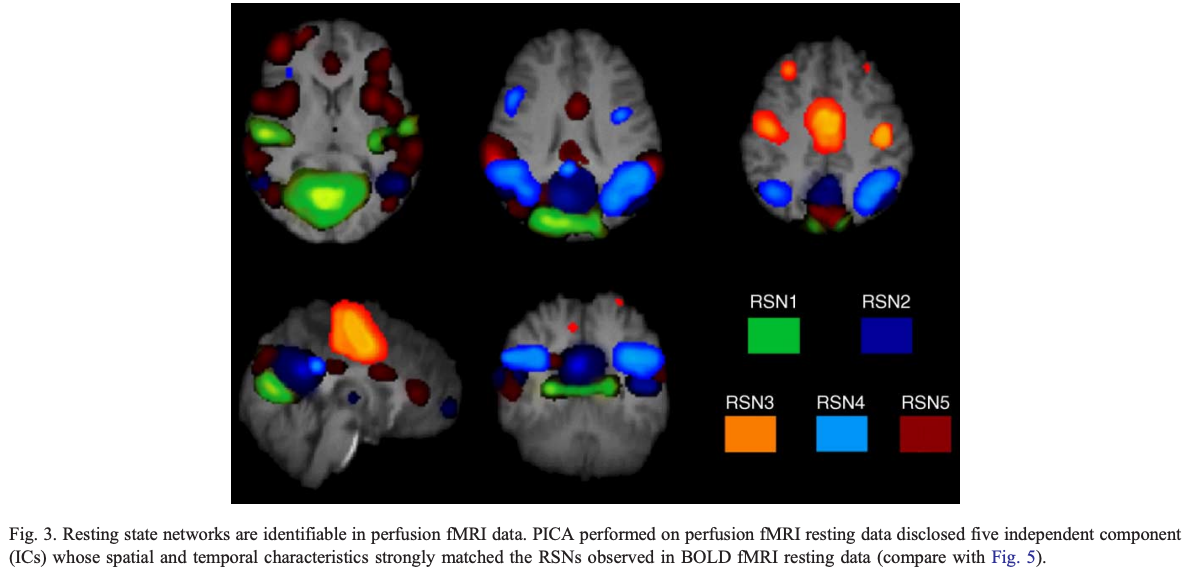
Early resting state networks De Luca 2006
Resting state networks (Damoiseaux)
Published the same year, using a different ICA method.
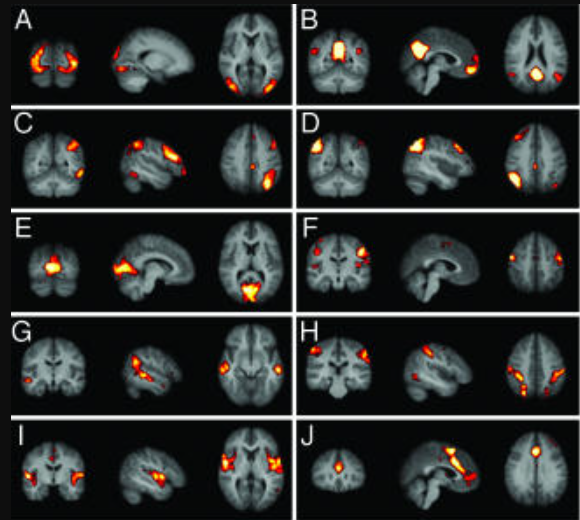
Early resting state networks Damoiseaux 2006
Diffusion imaging
Measures features of water “diffusion” in the brain. Typically, to estimate biological features of the white matter
- Will not focus on this data type in this course
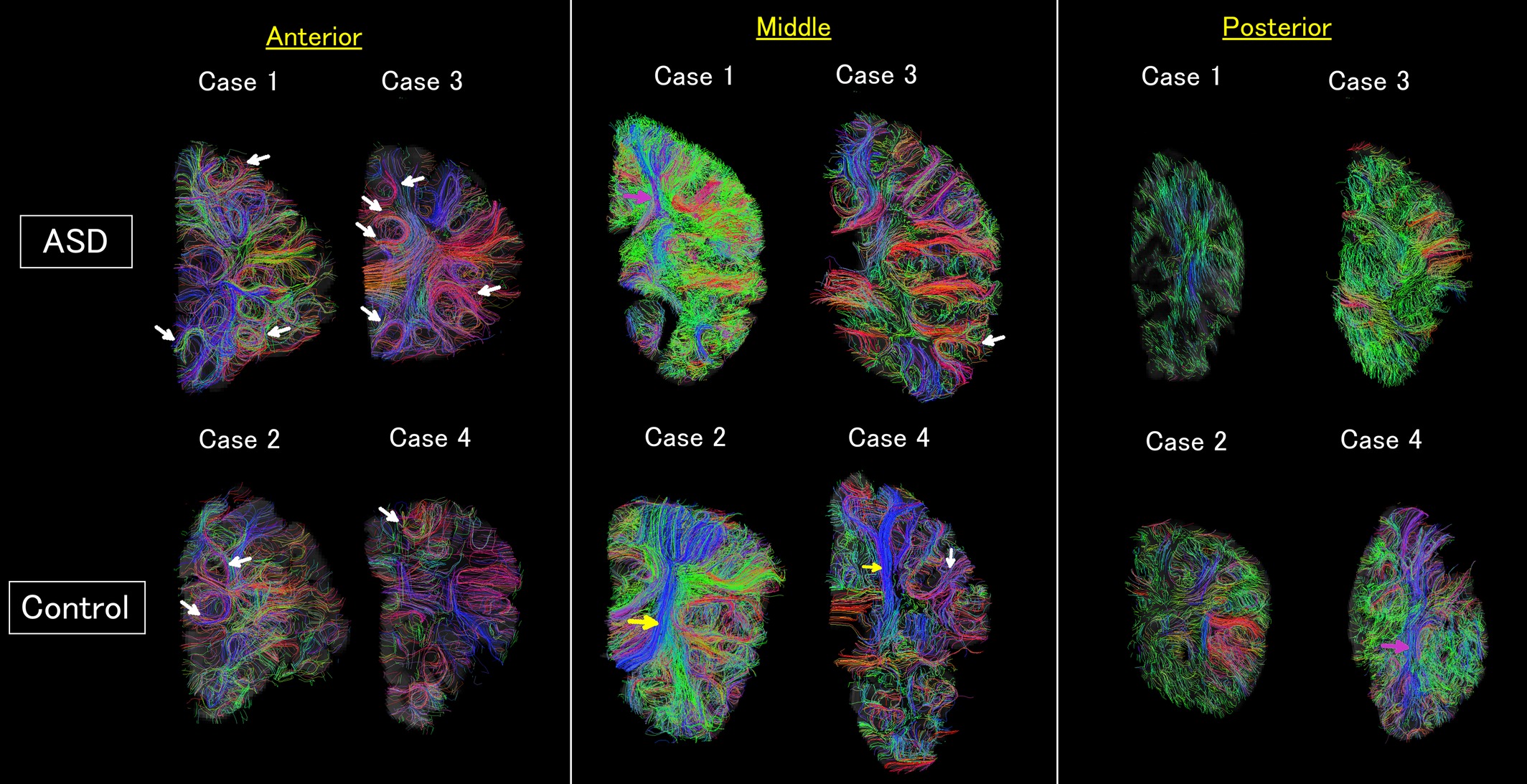
Diffusion imaging in Autism Wilkinson 2016
Other measures
- Perfusion – Quantitative measure of blood flow
- Spectroscopy
- Many others actively in development
- Many “derivatives” exist and in development
- Derivatives are quantitative values derived from existing modalities