First-level Analysis
Objectives
- These slides will introduce first-level fMRI analysis
- Part 1: task fMRI
- Part 2: time series analysis
- Part 3: resting state fMRI
What is fMRI?
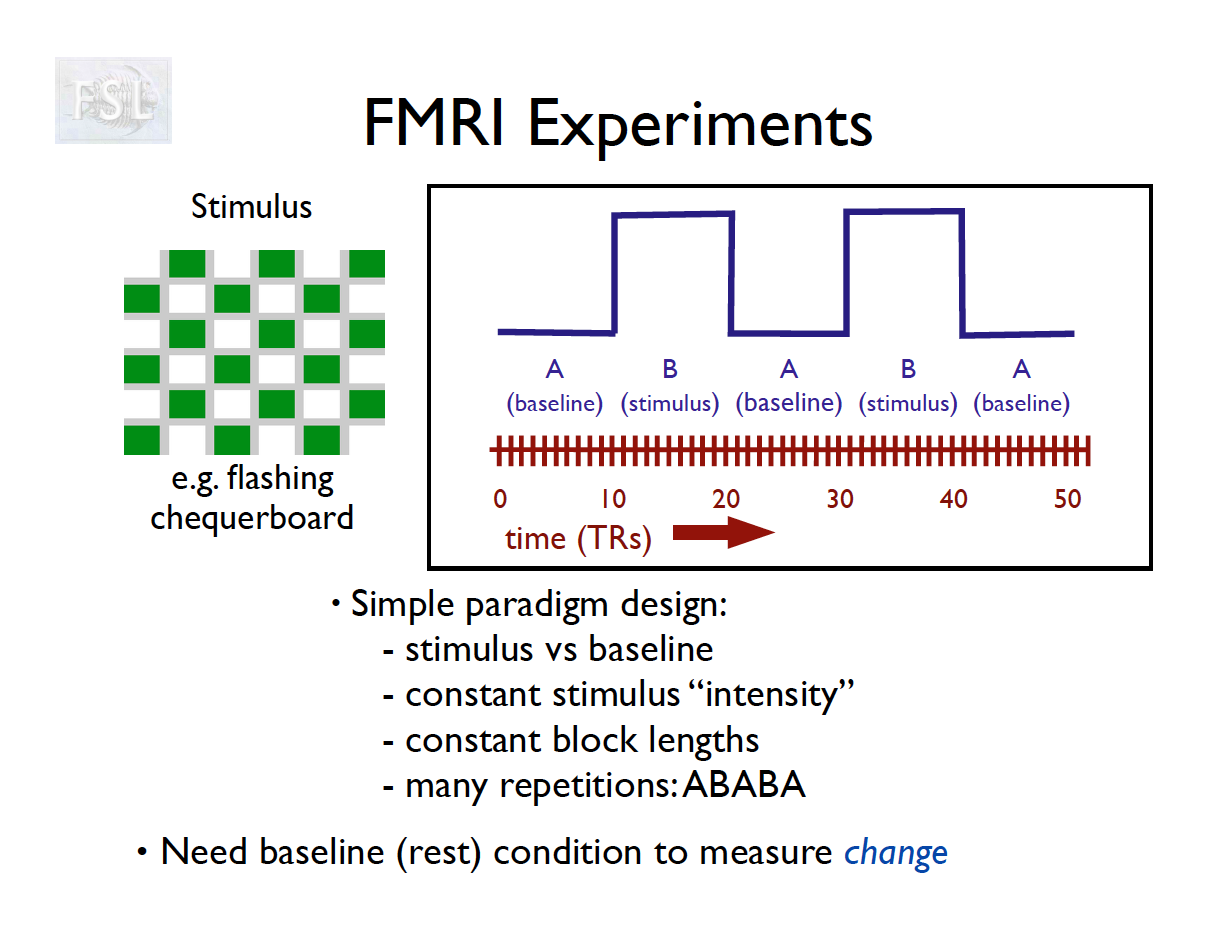
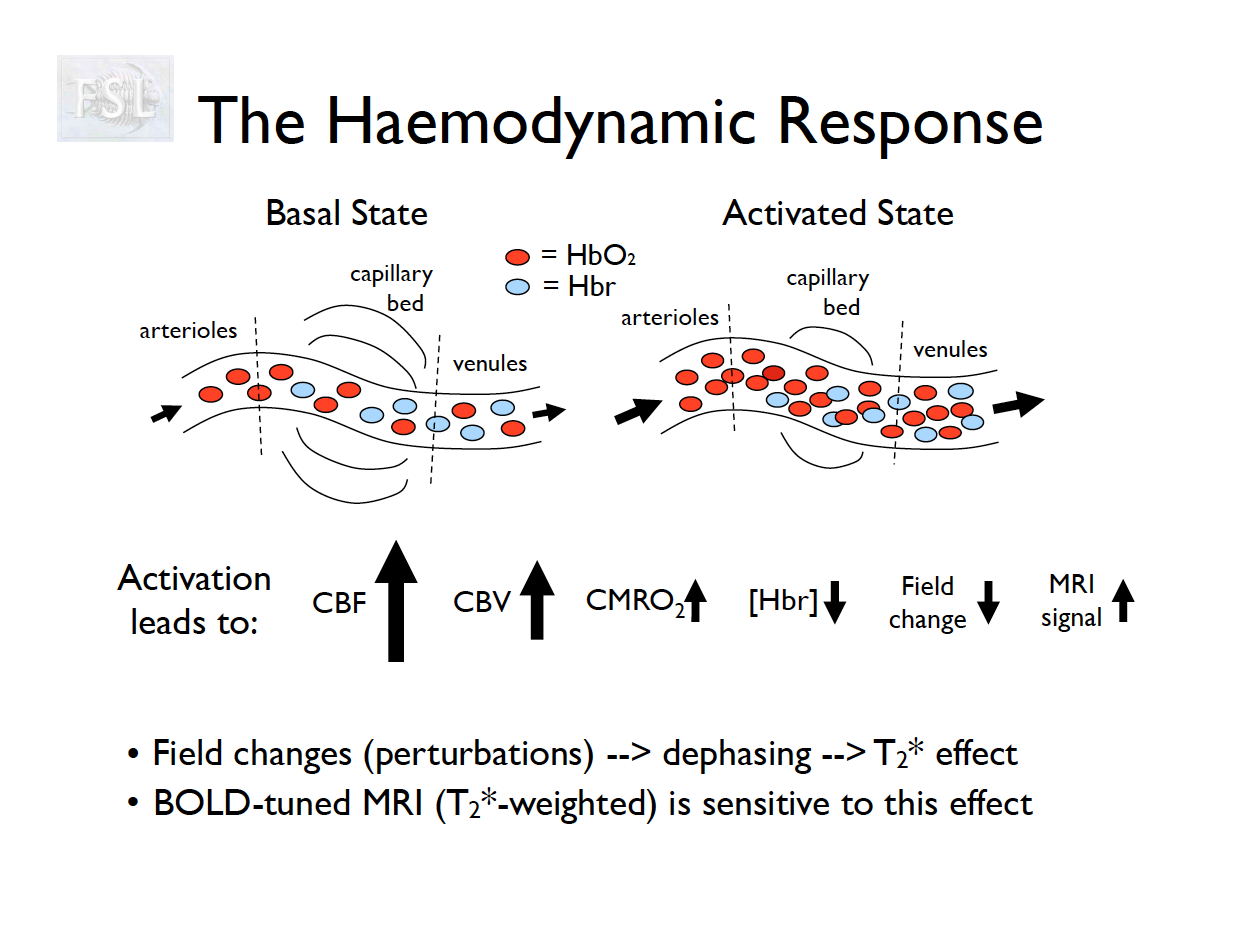
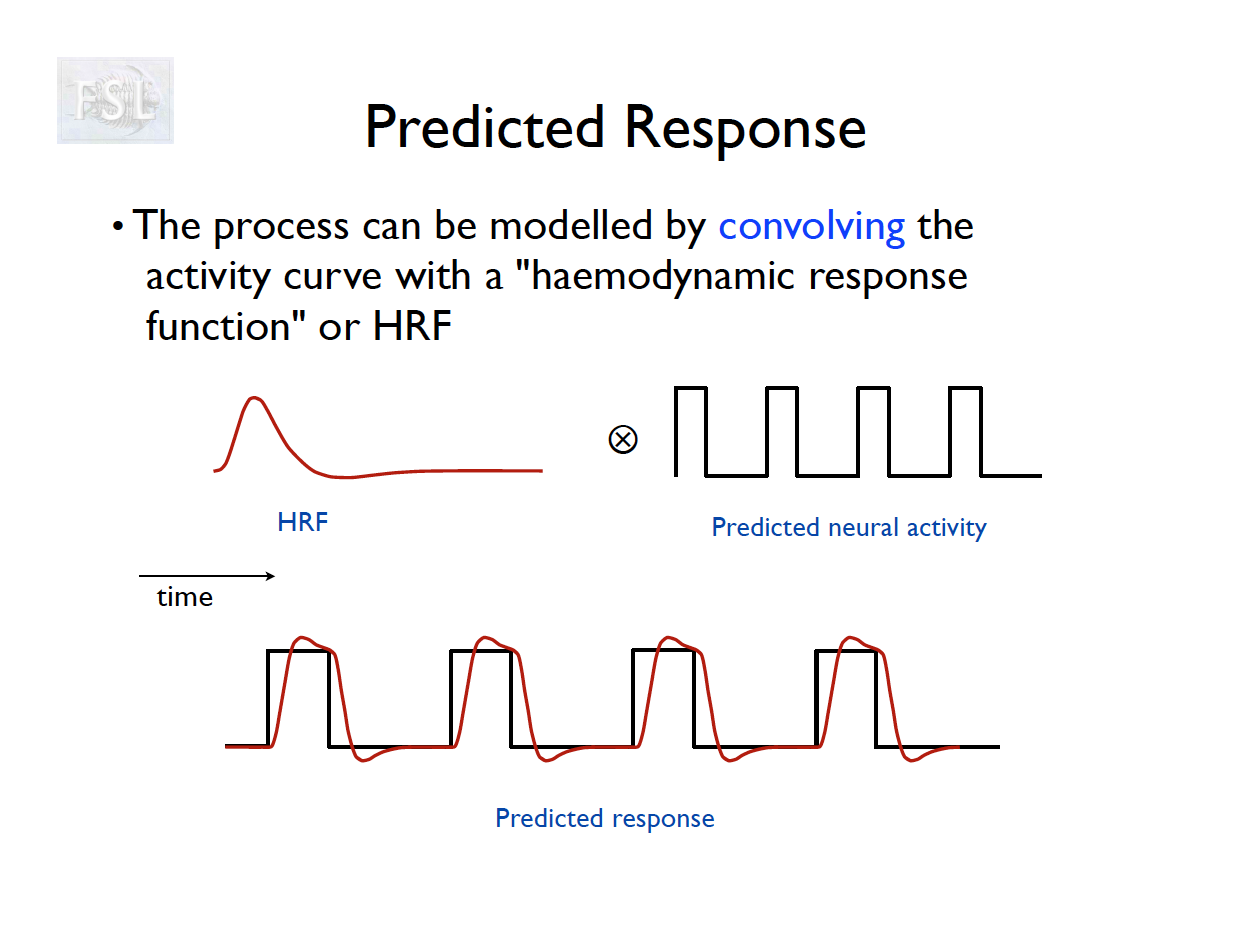
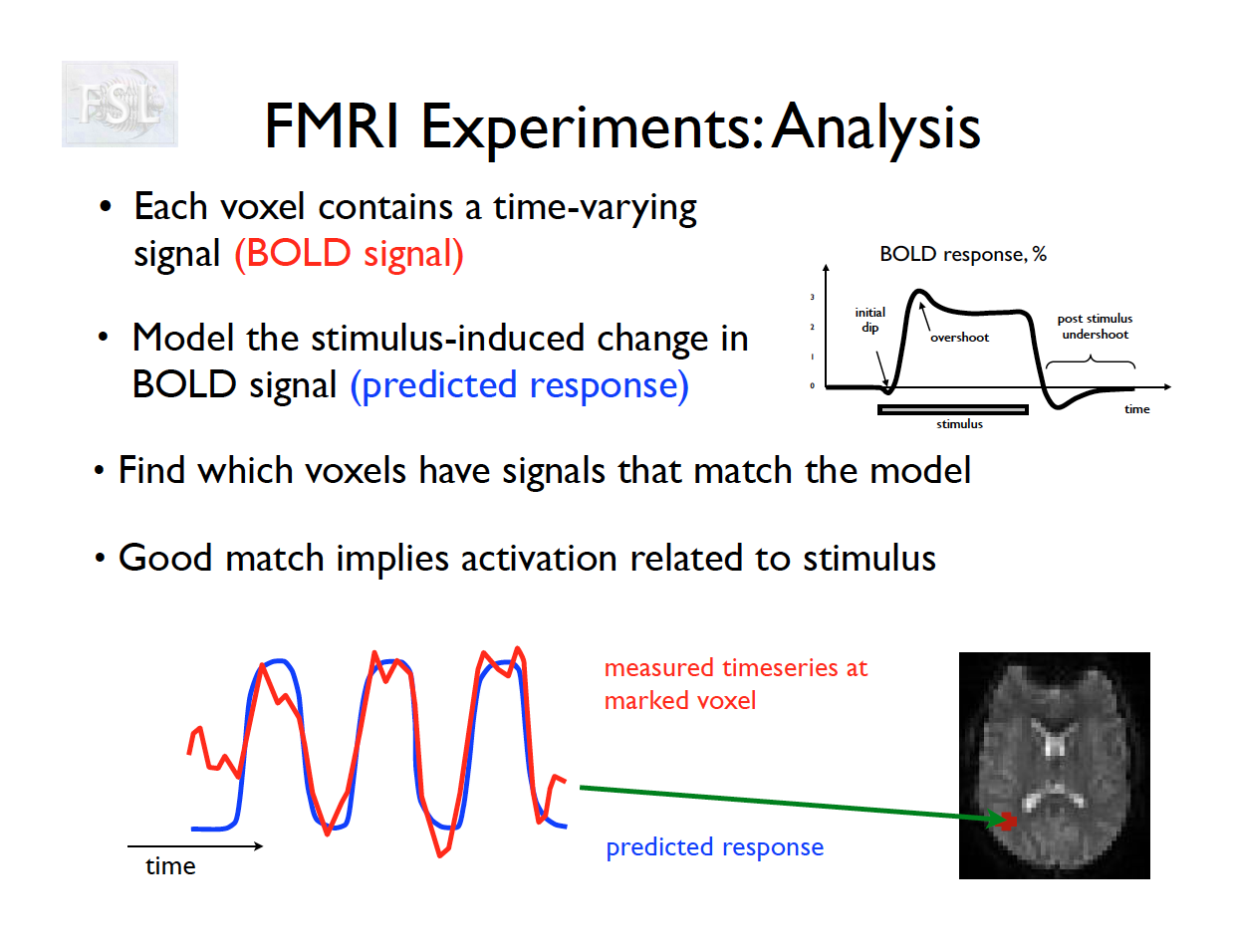
Basic task fMRI Analysis
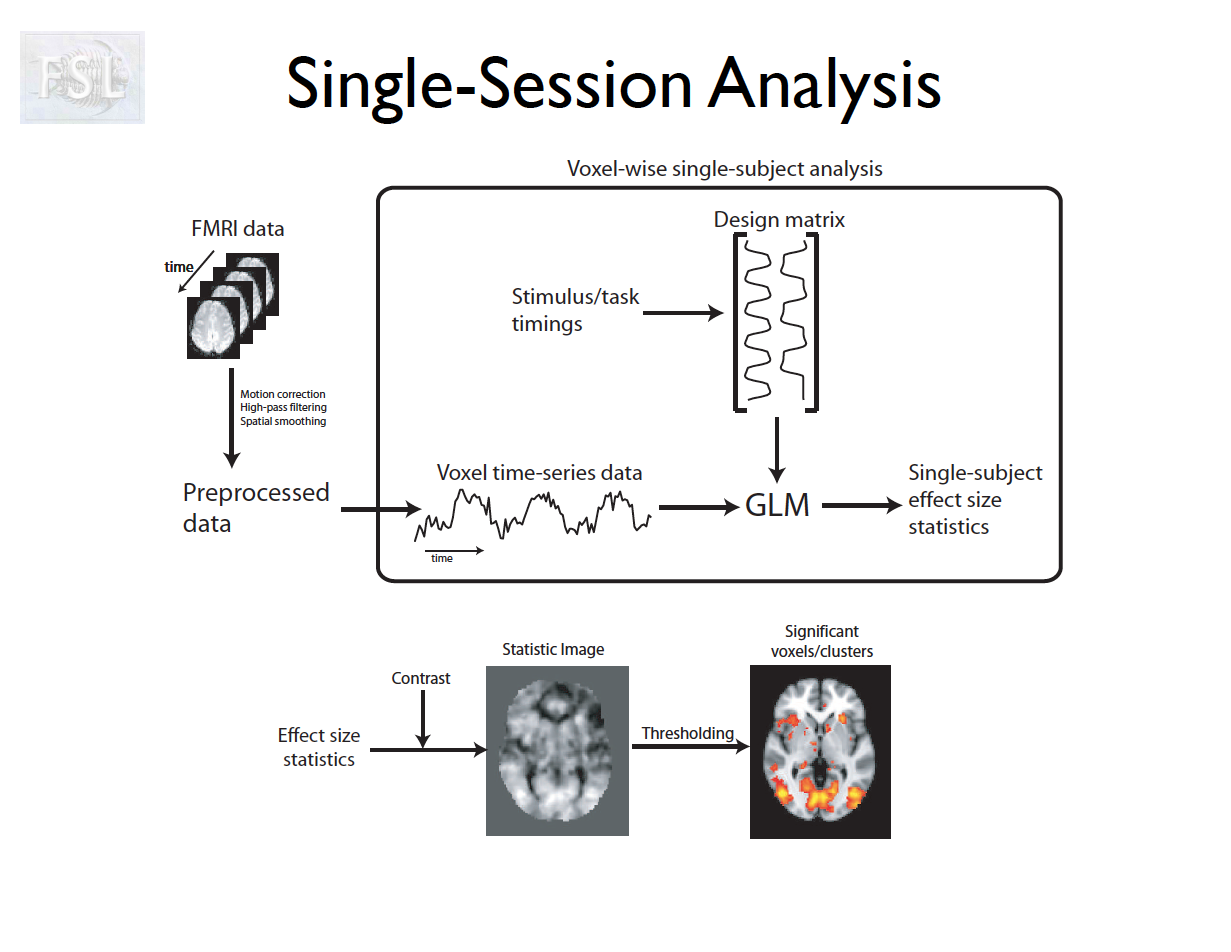
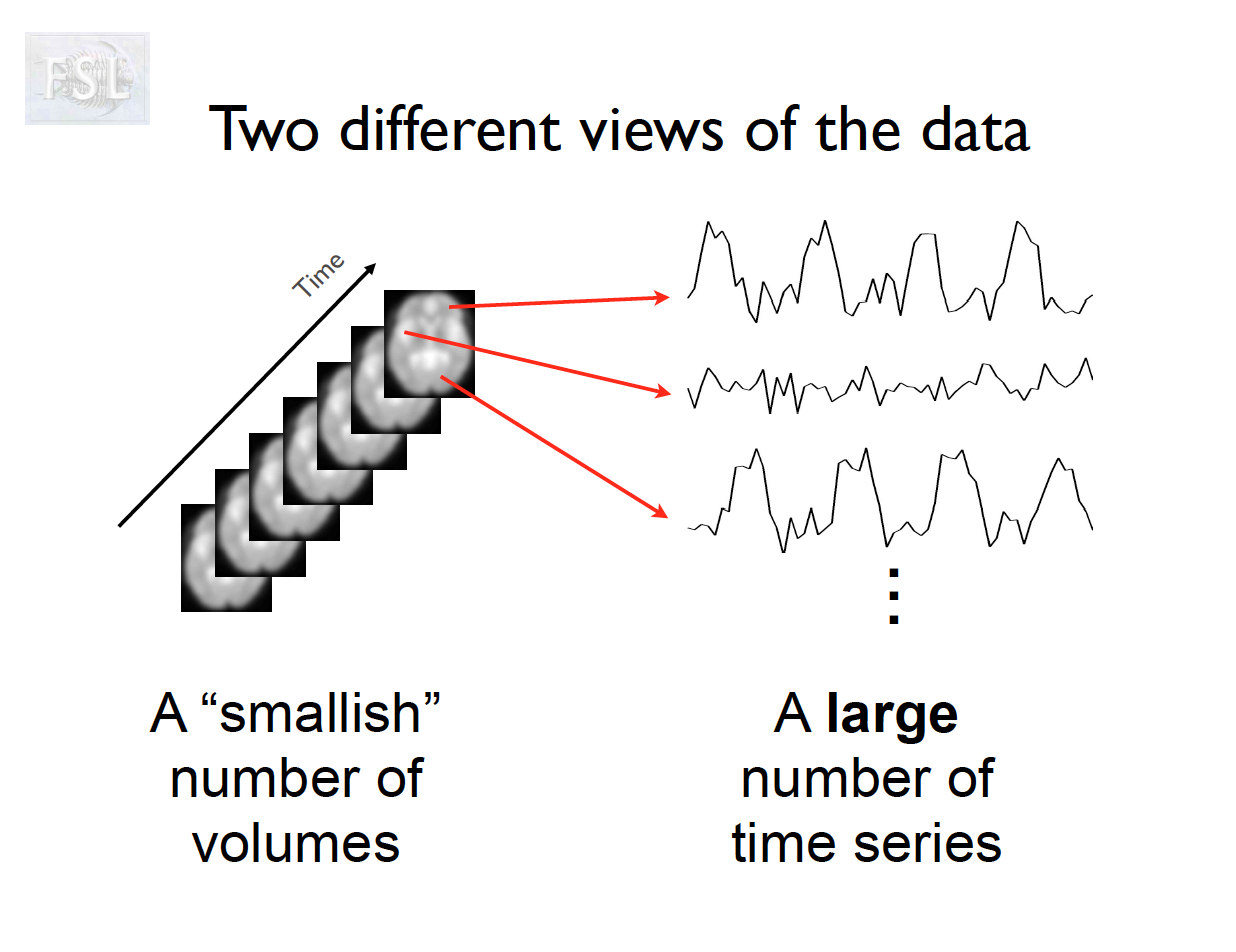
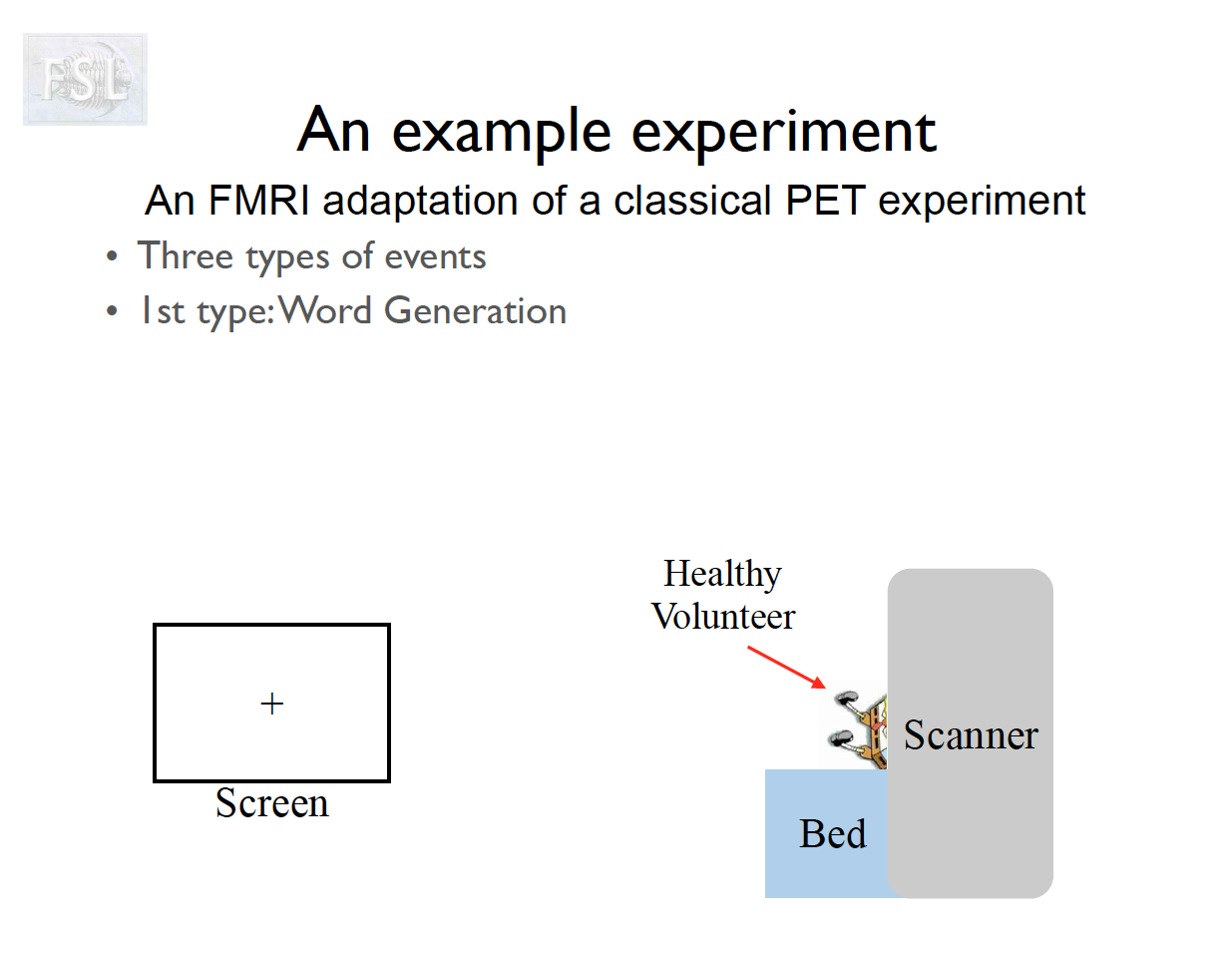
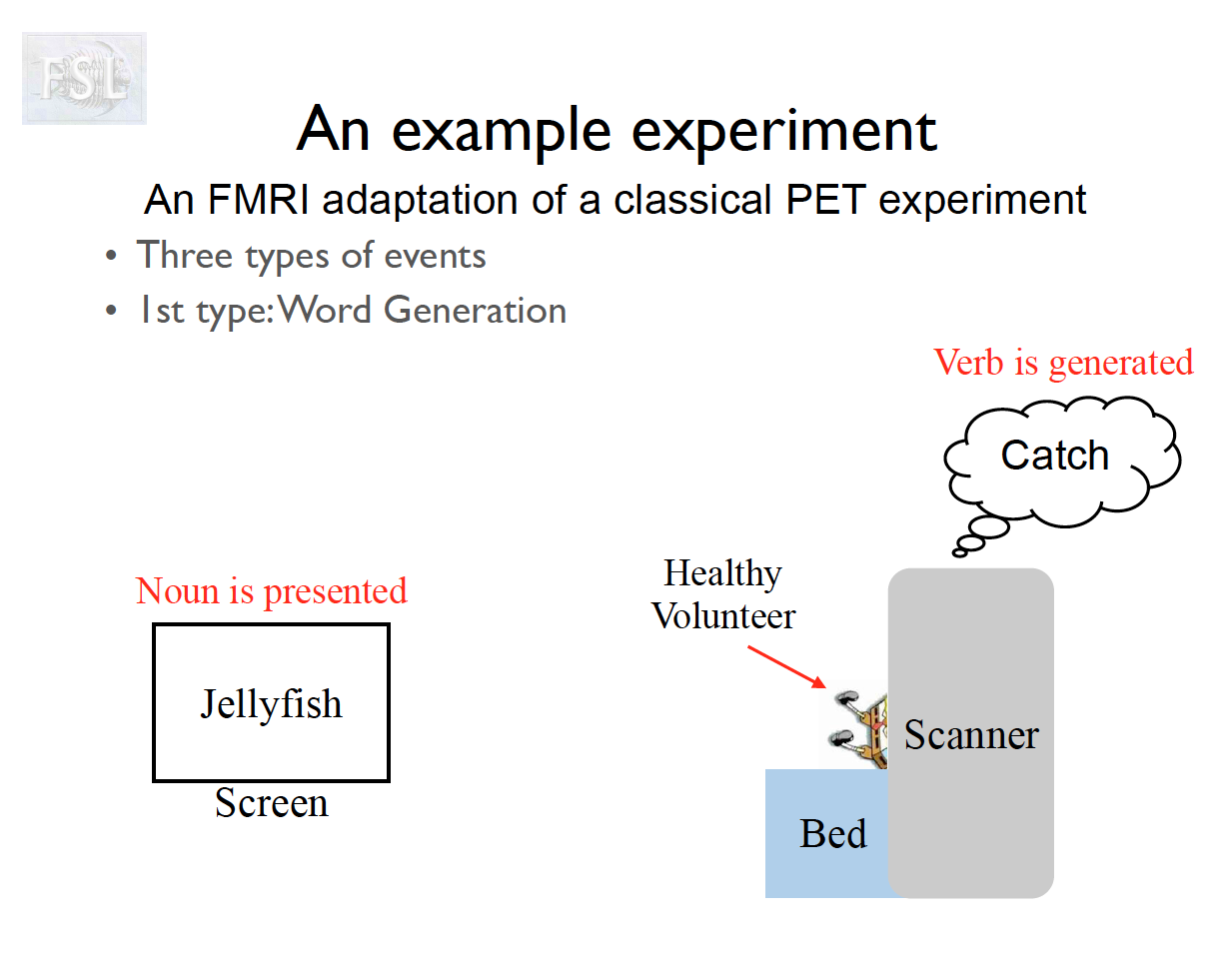
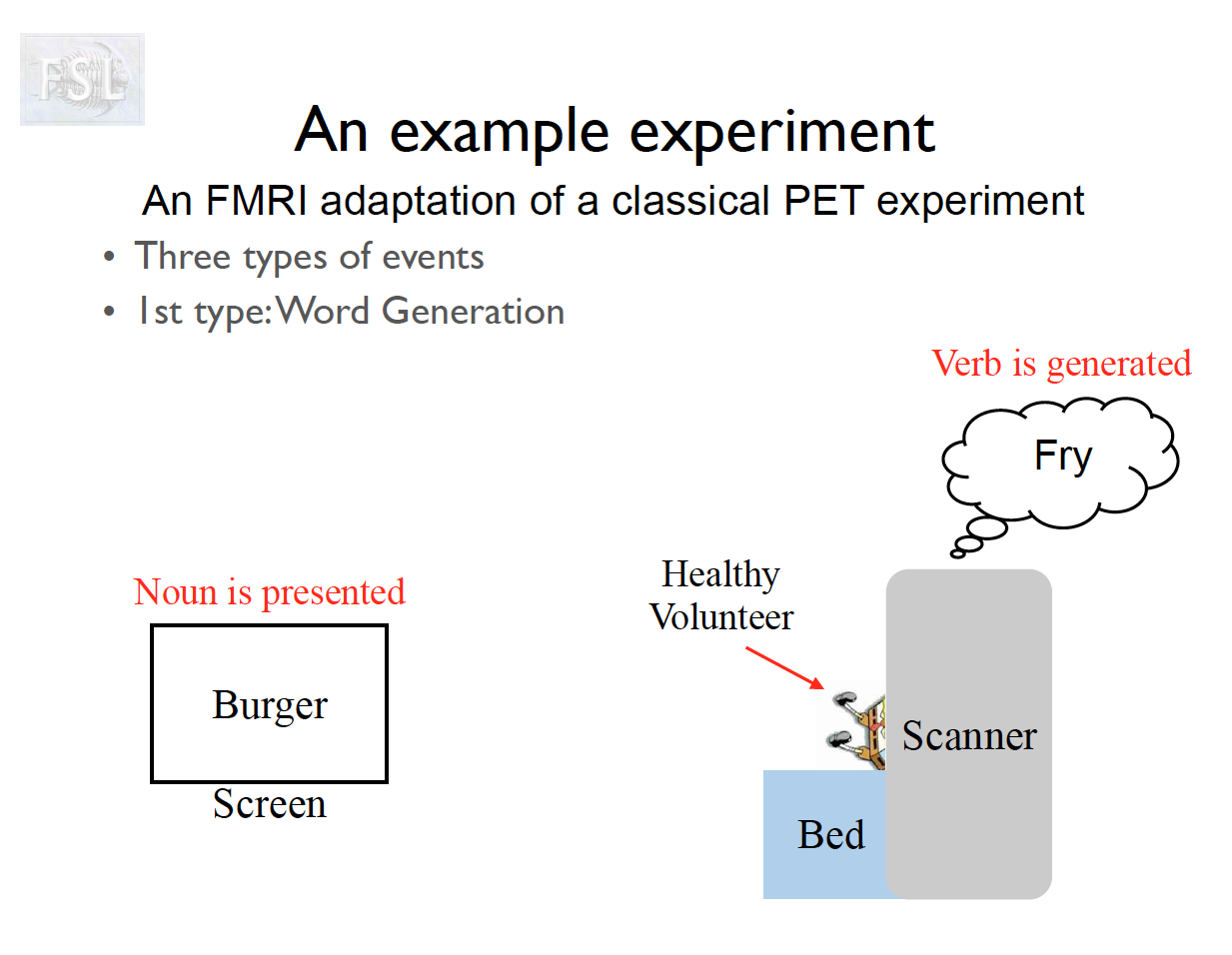
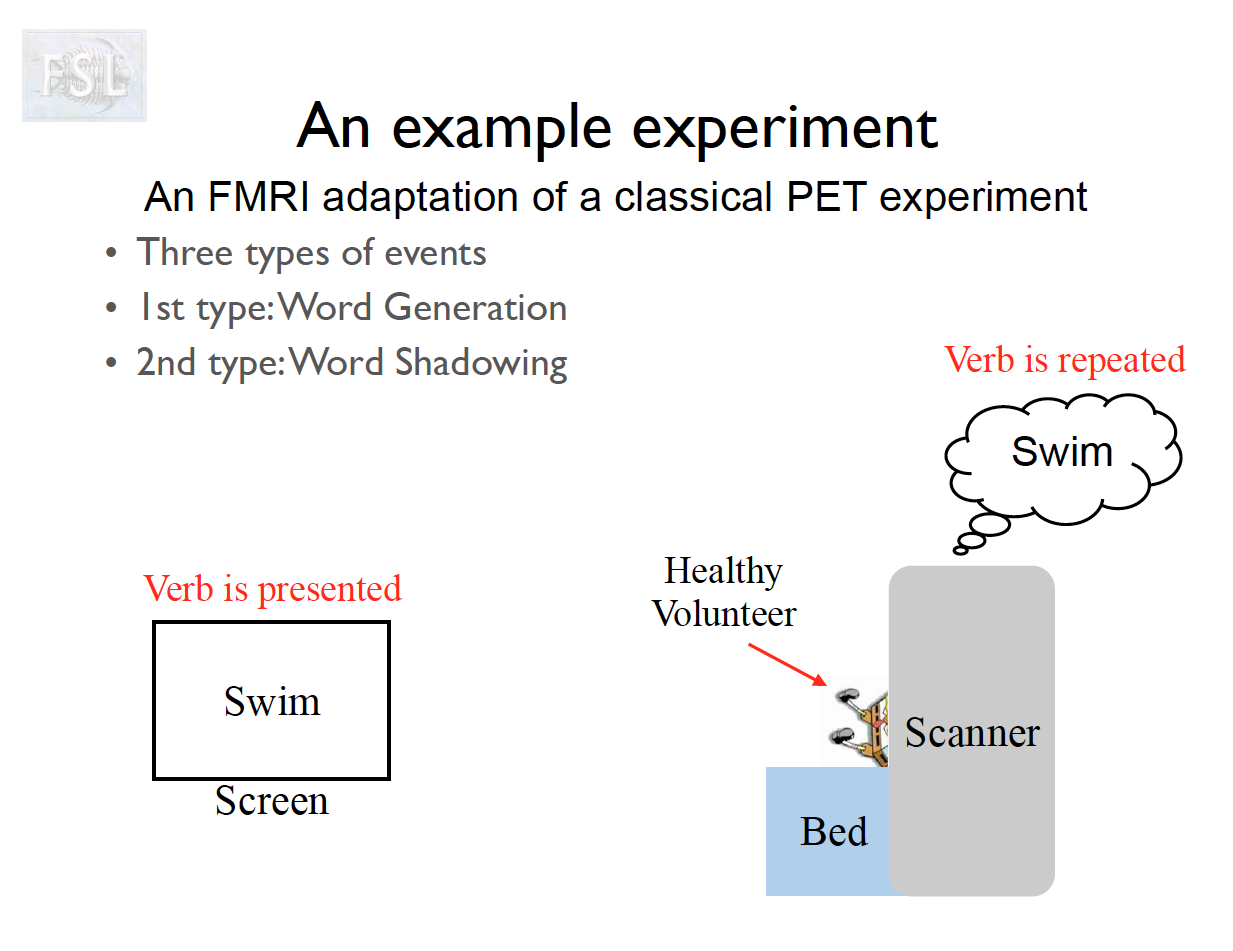
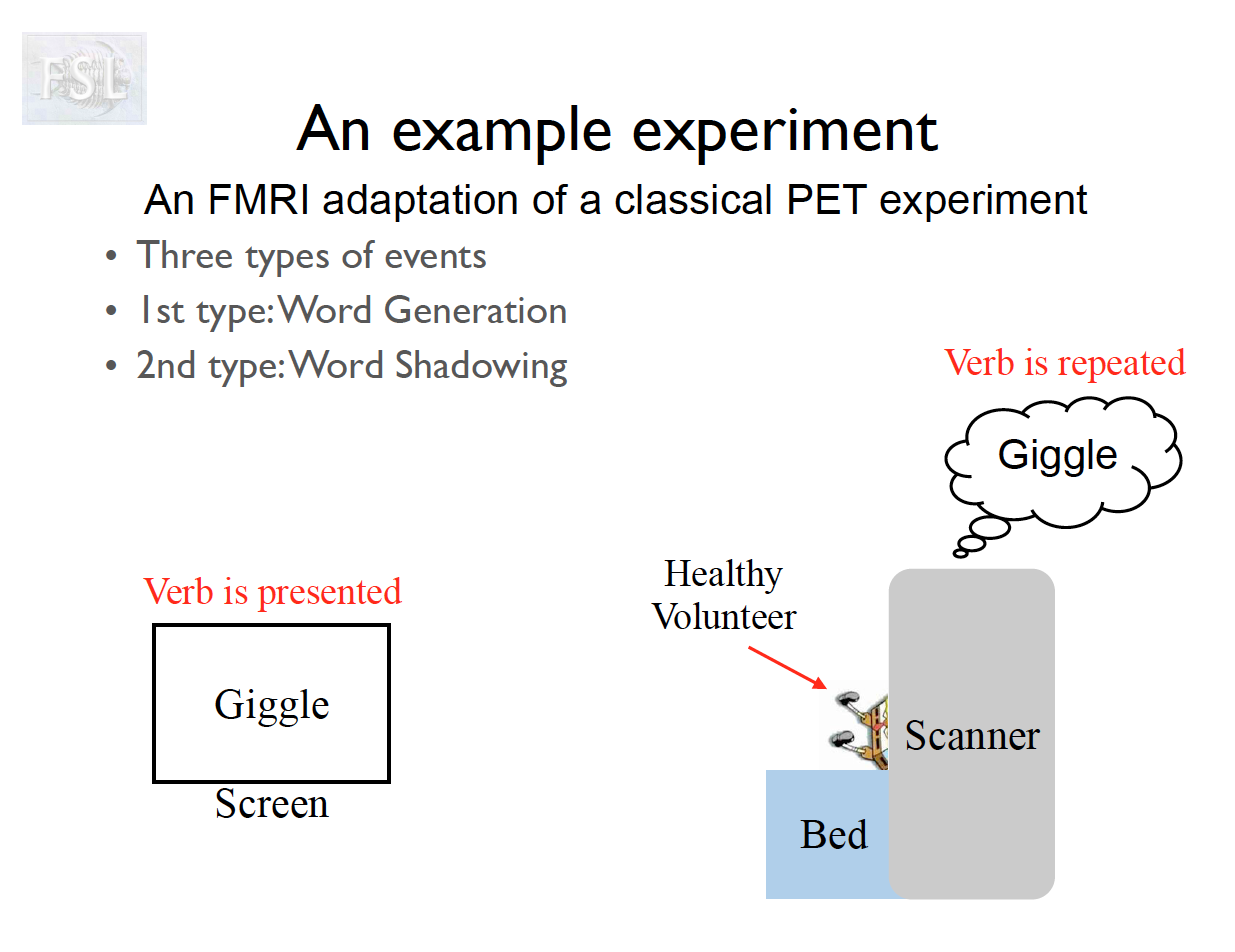
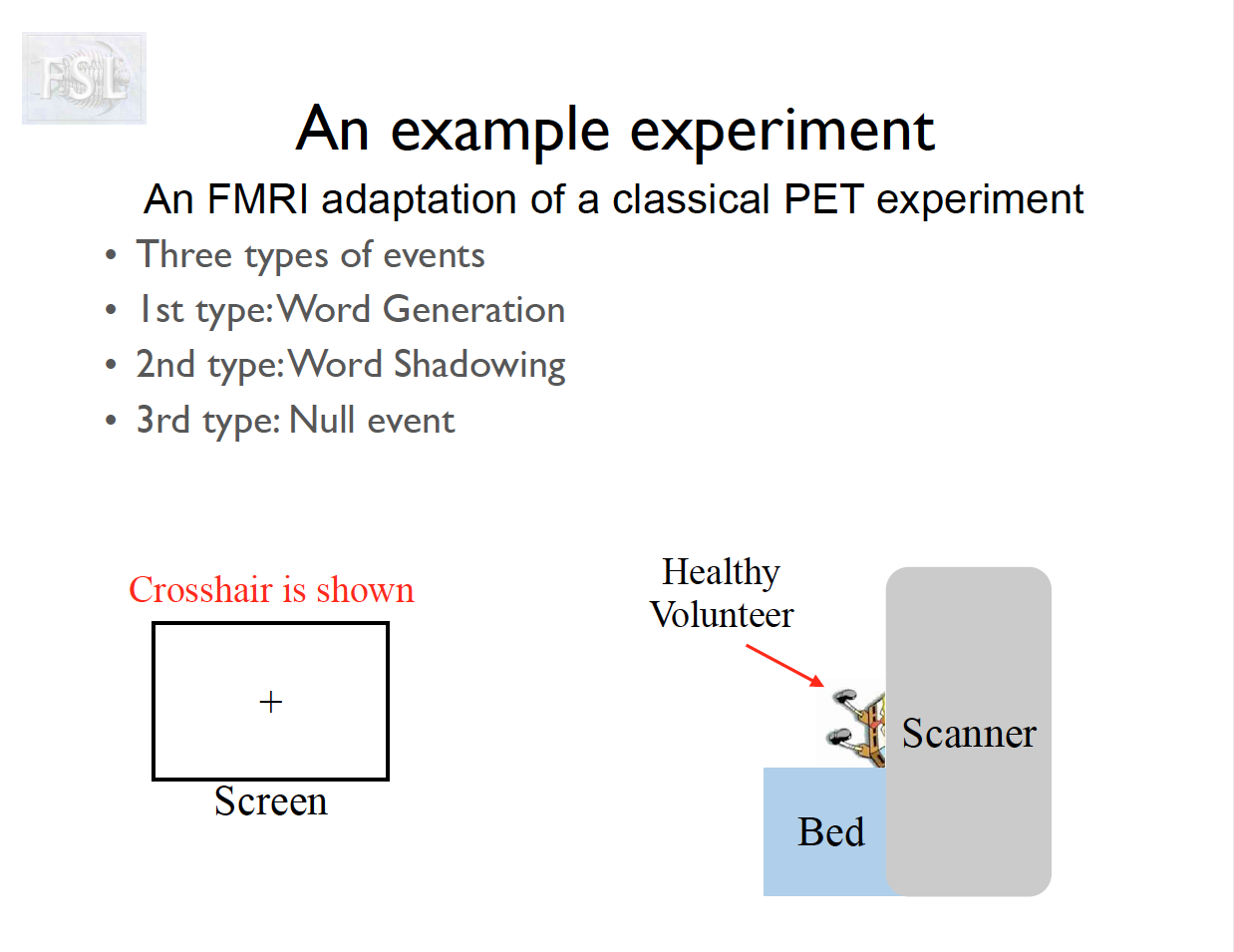
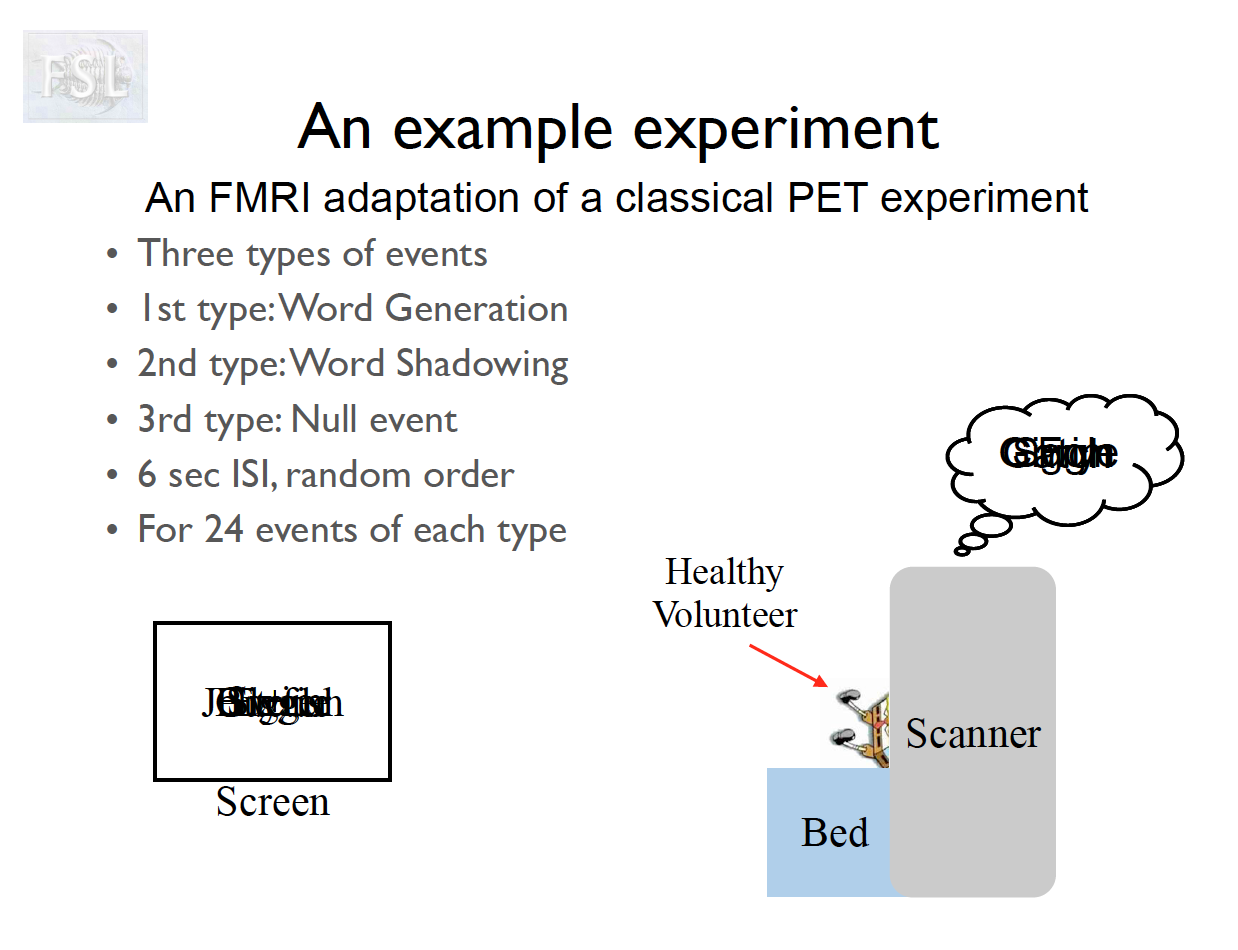
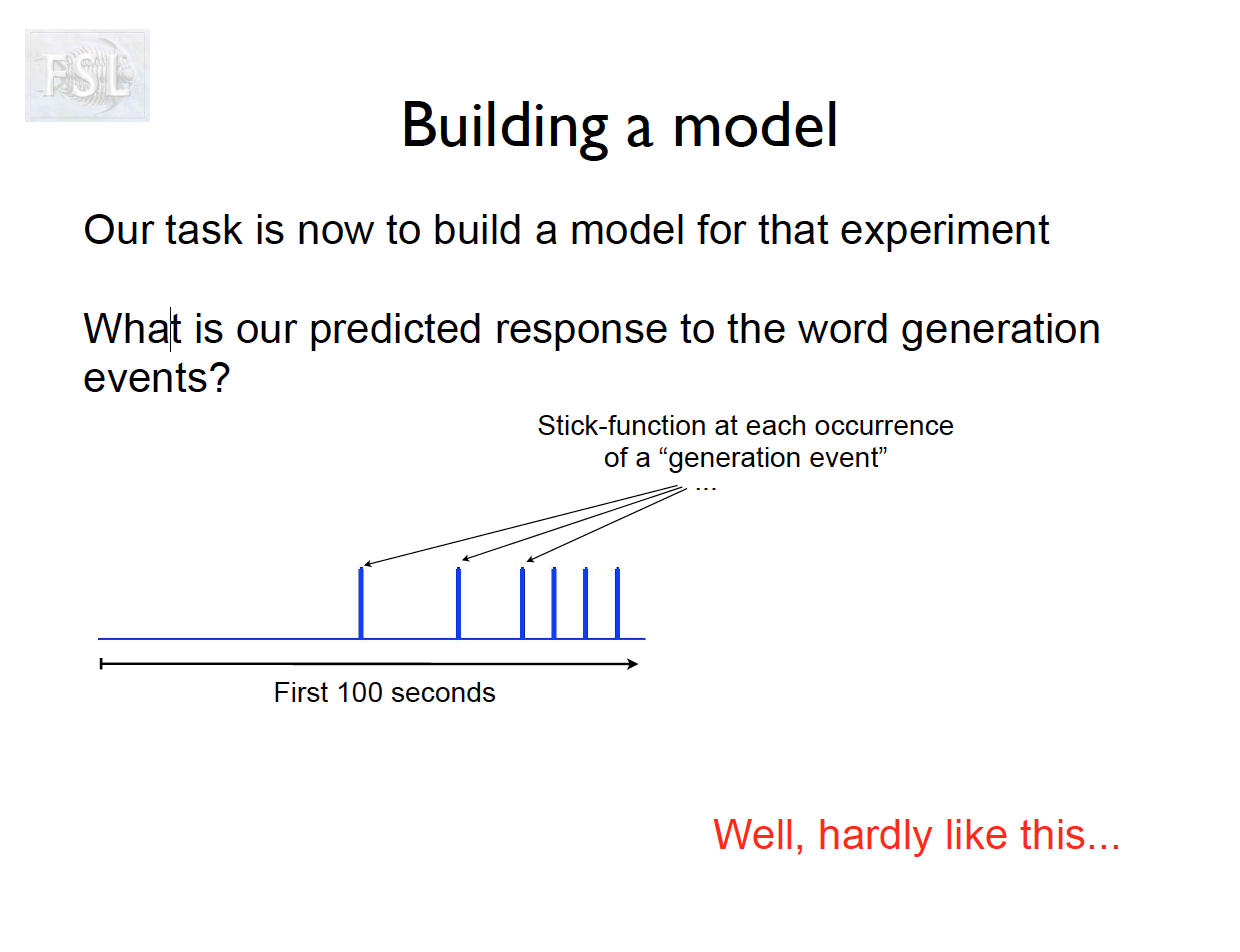
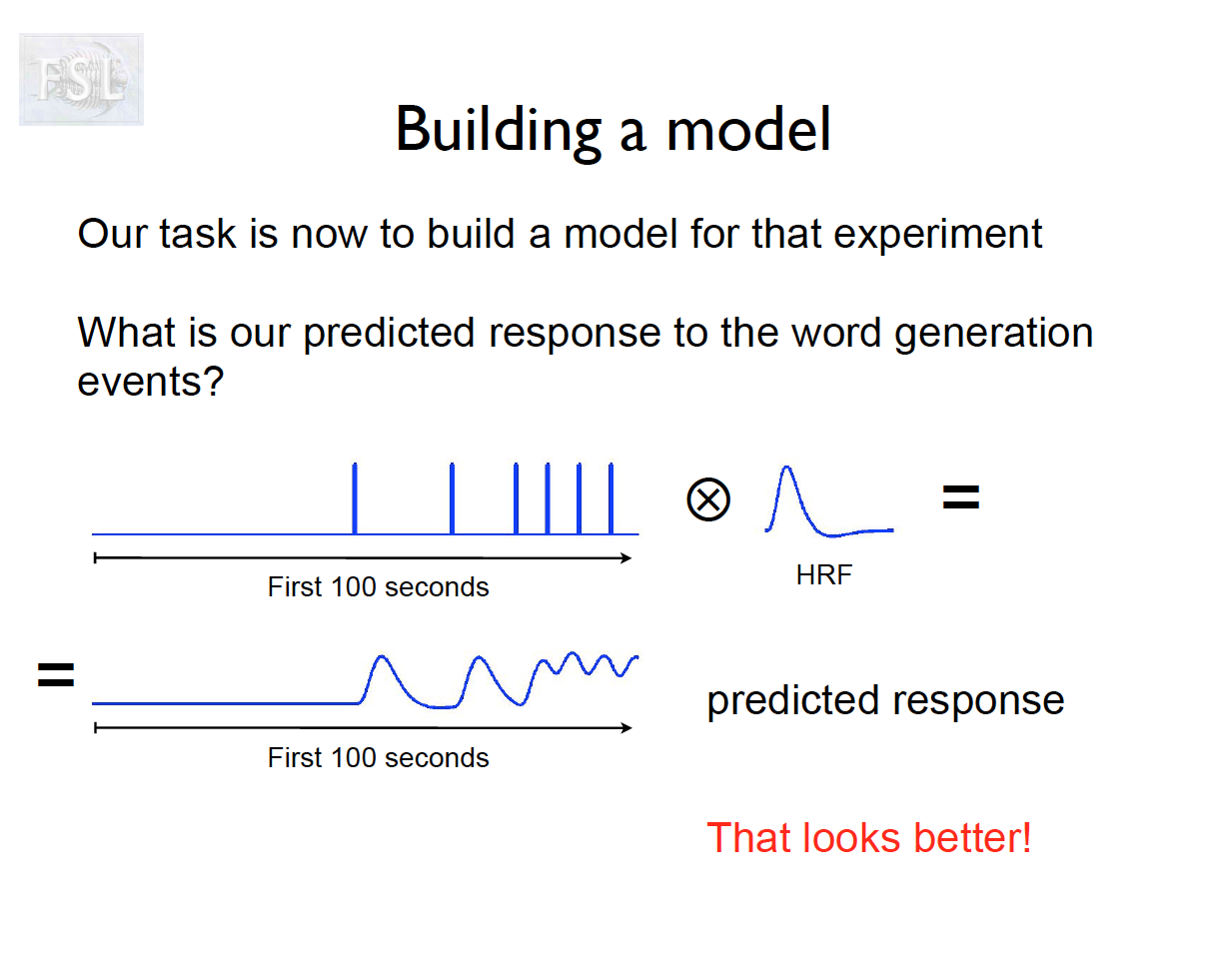
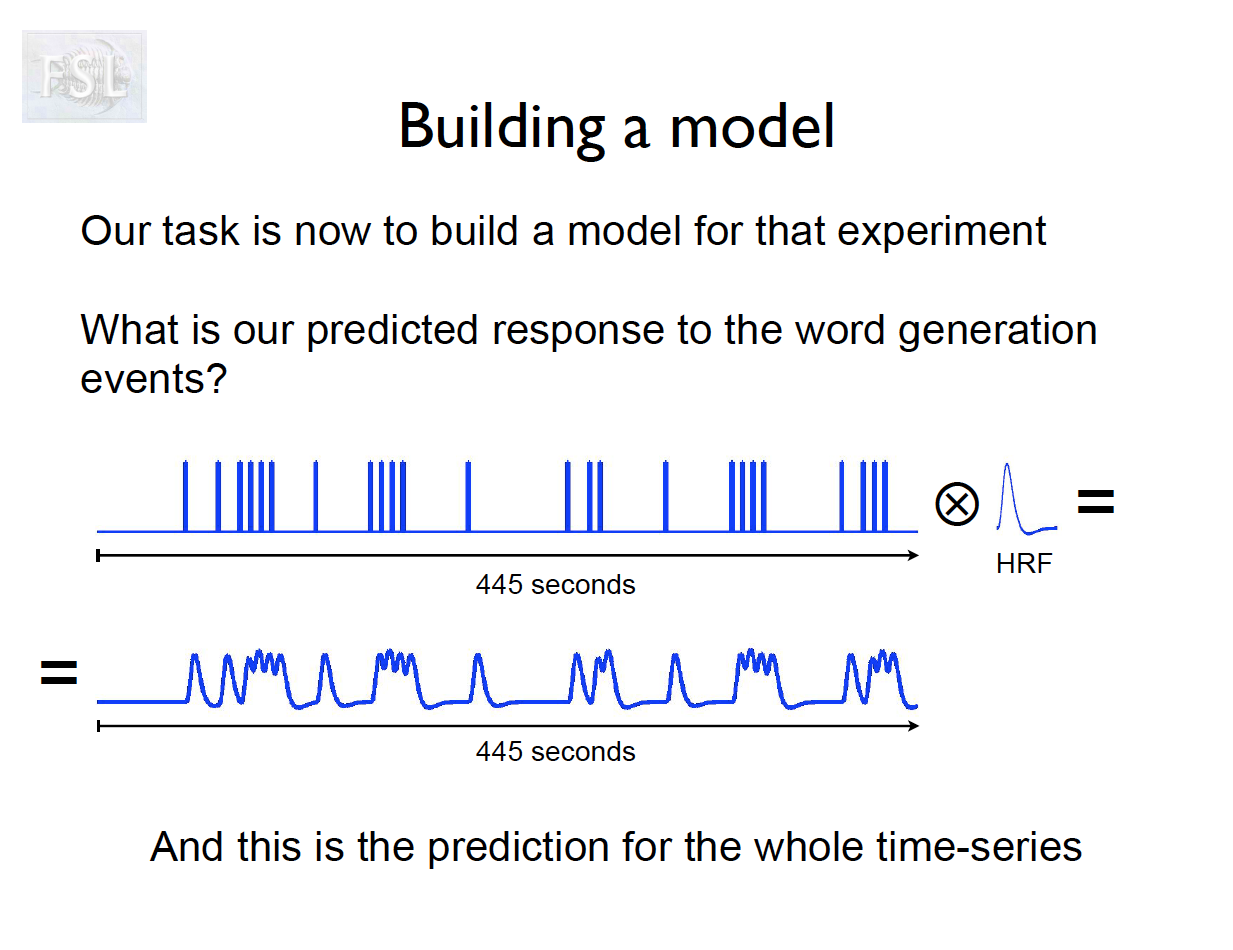
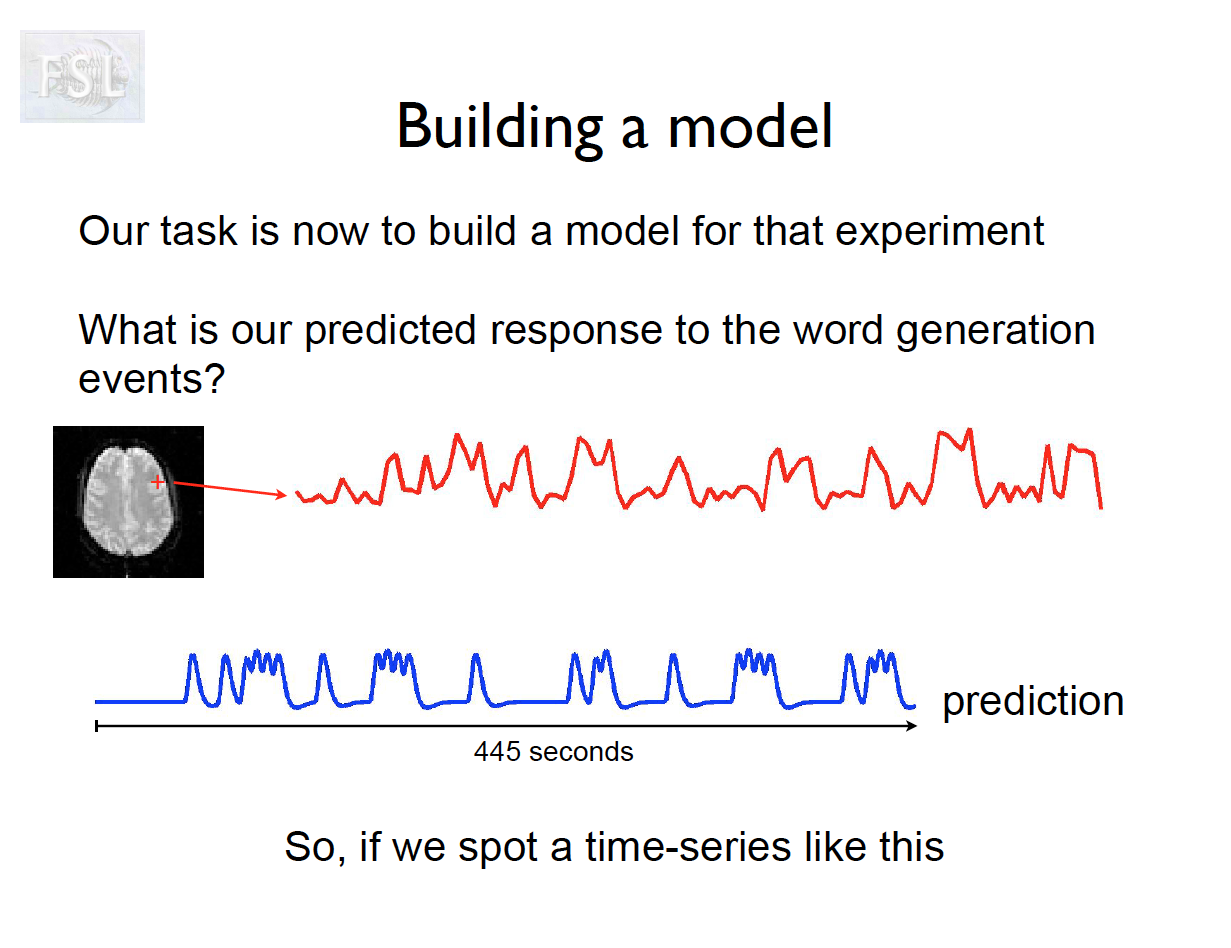
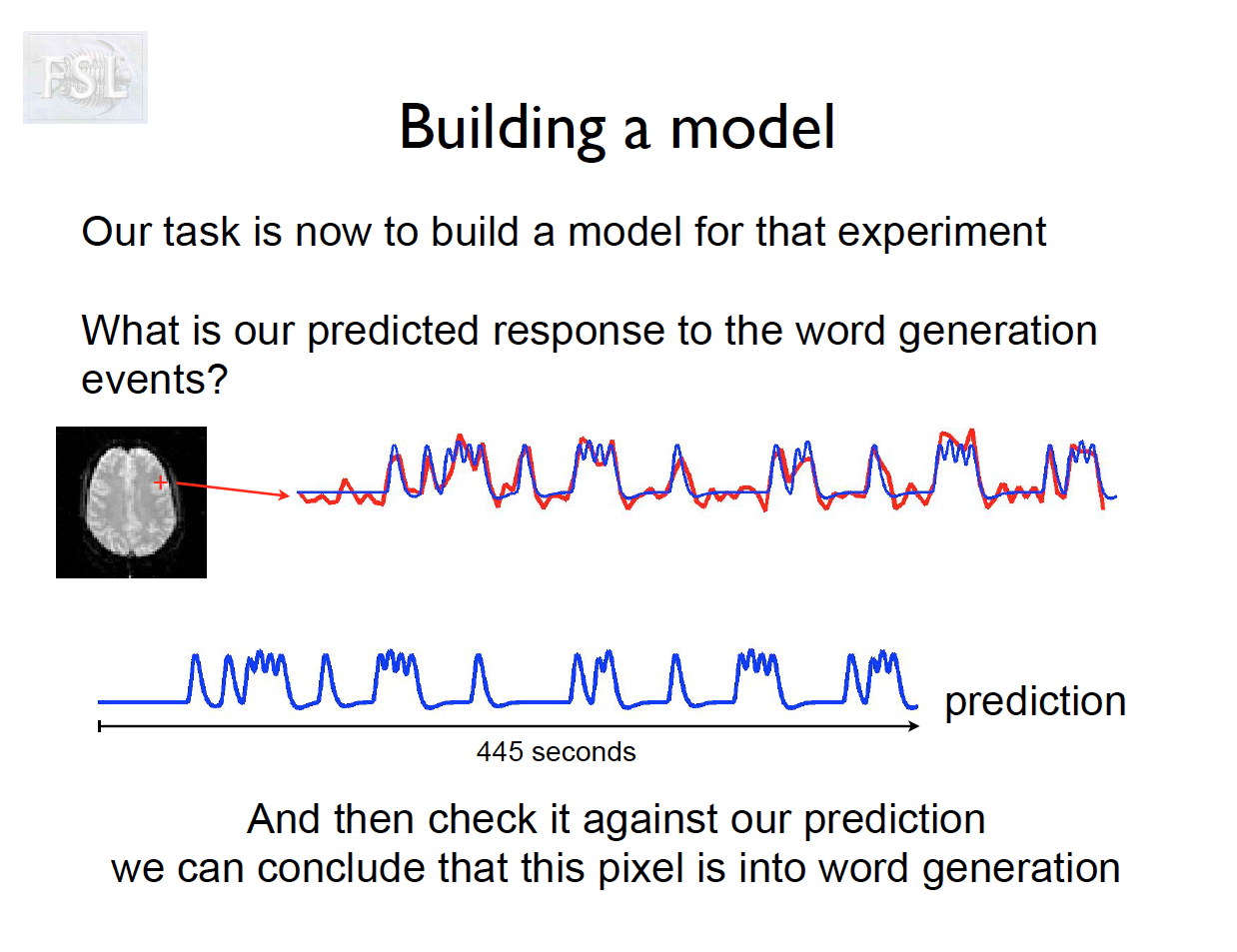

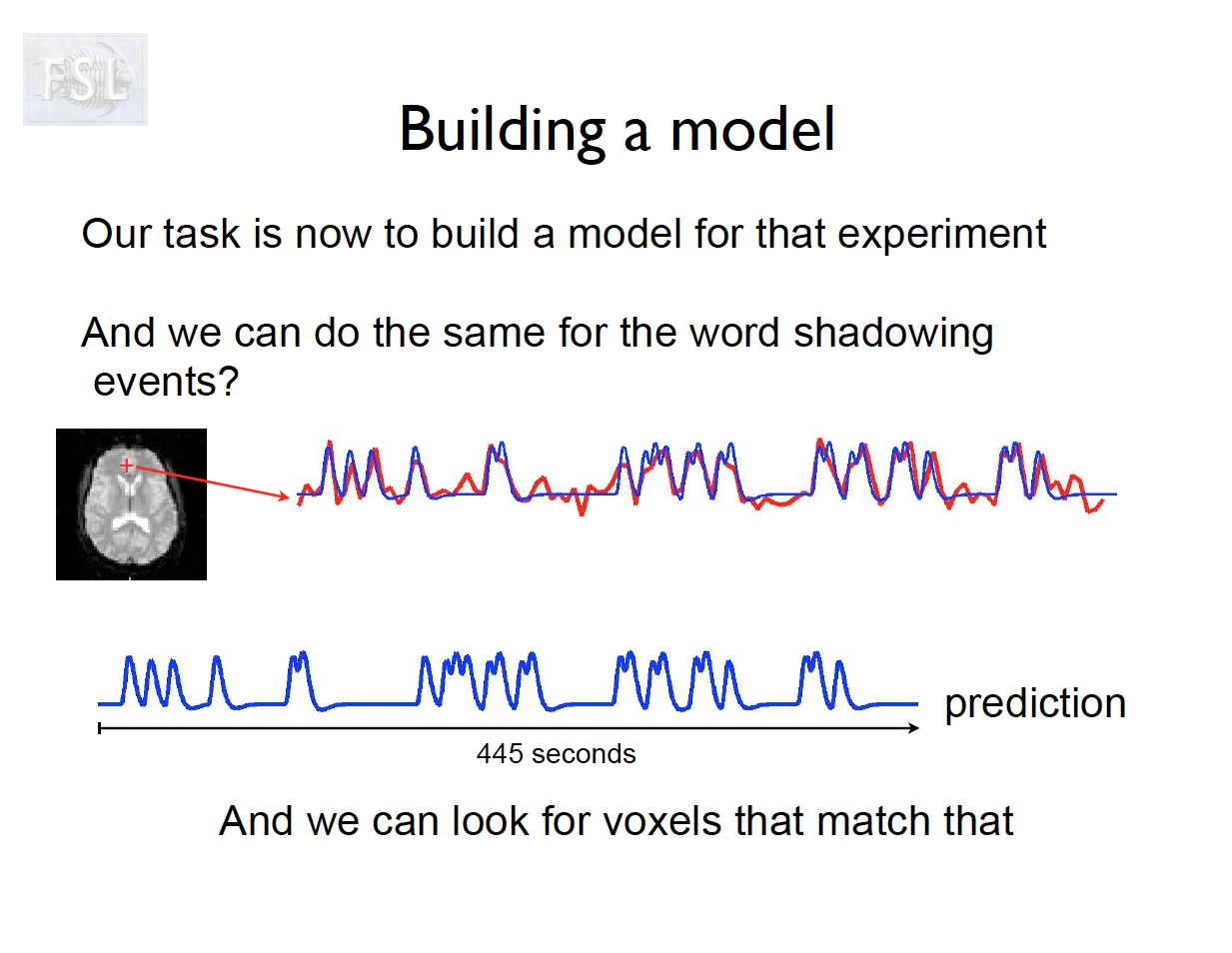
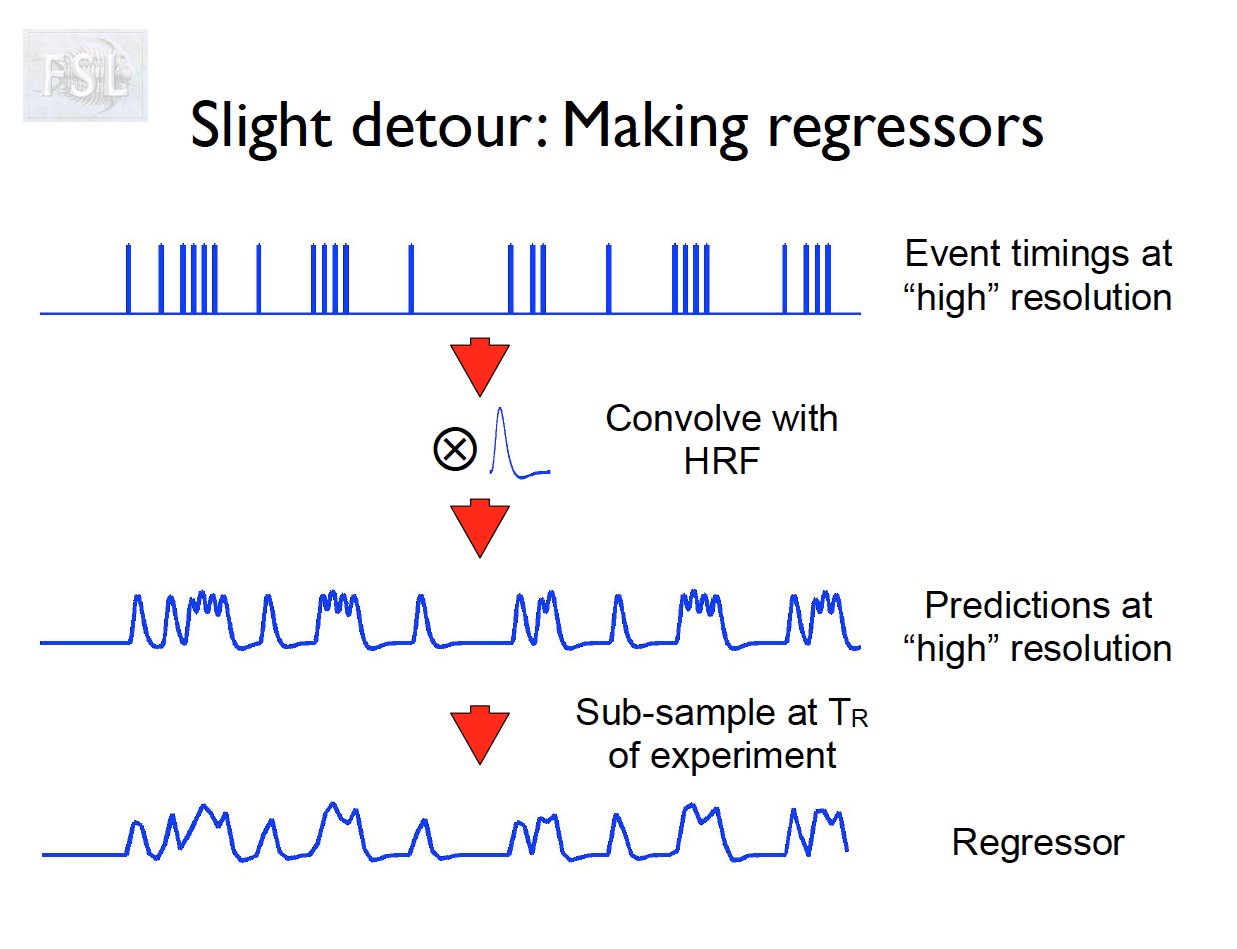
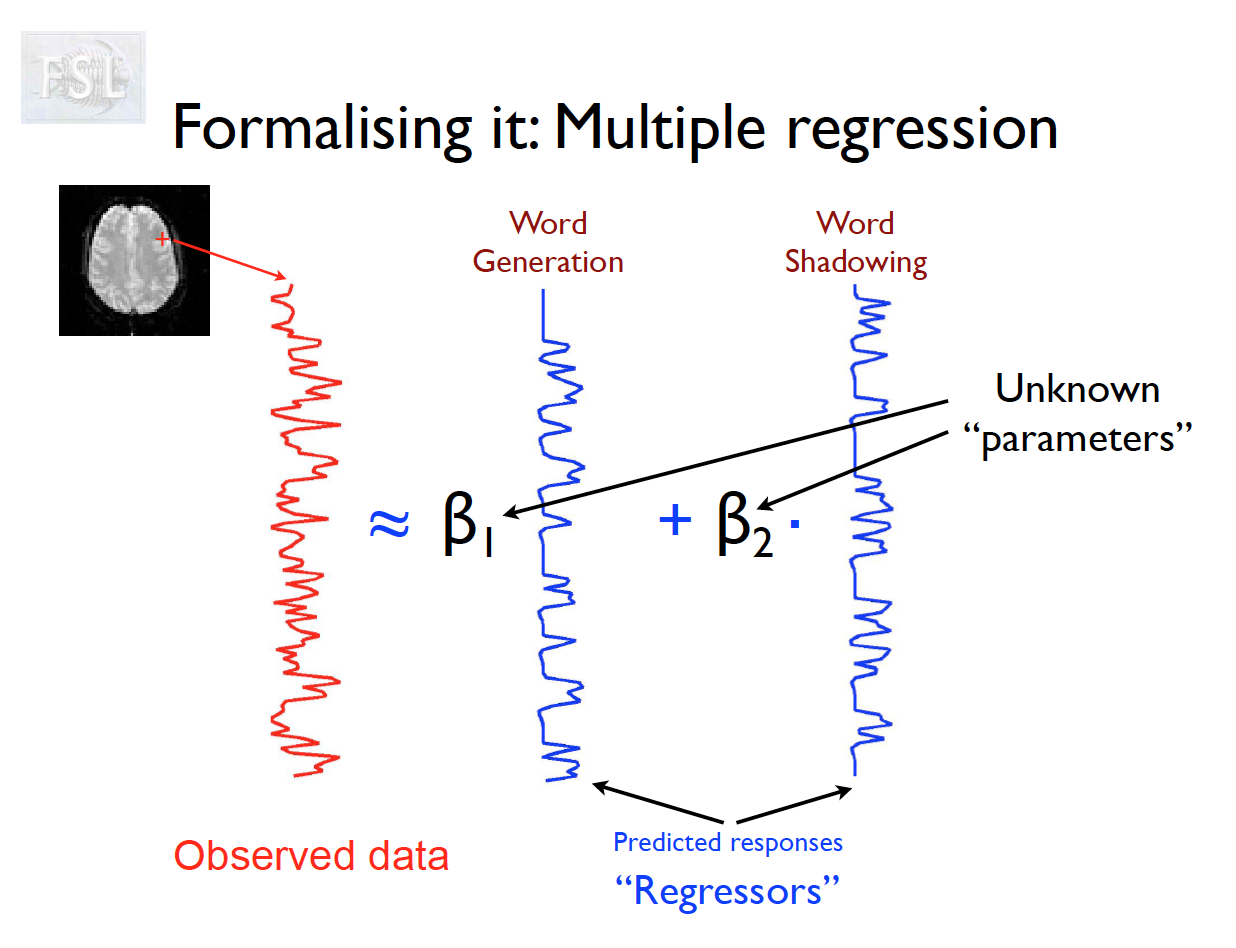
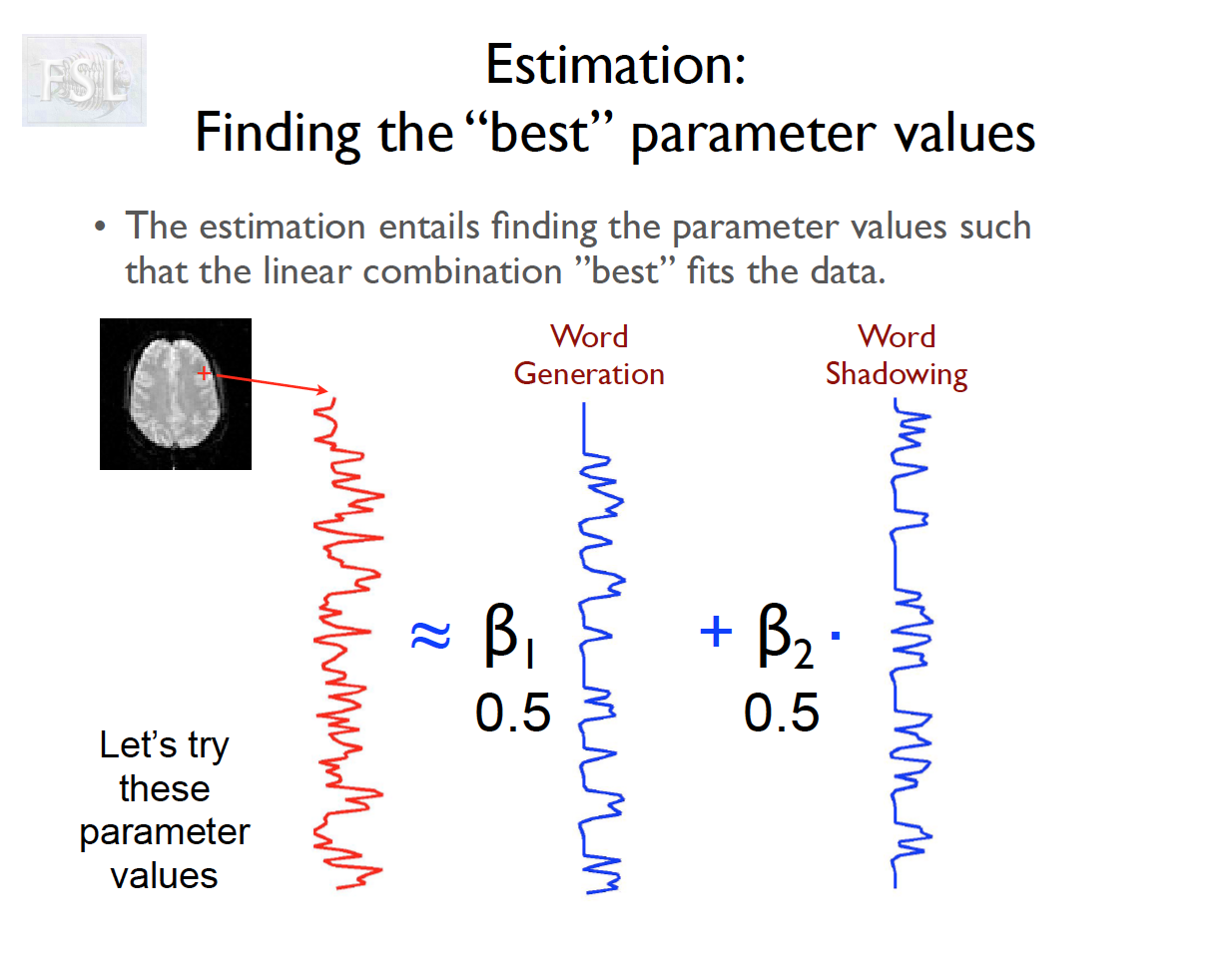
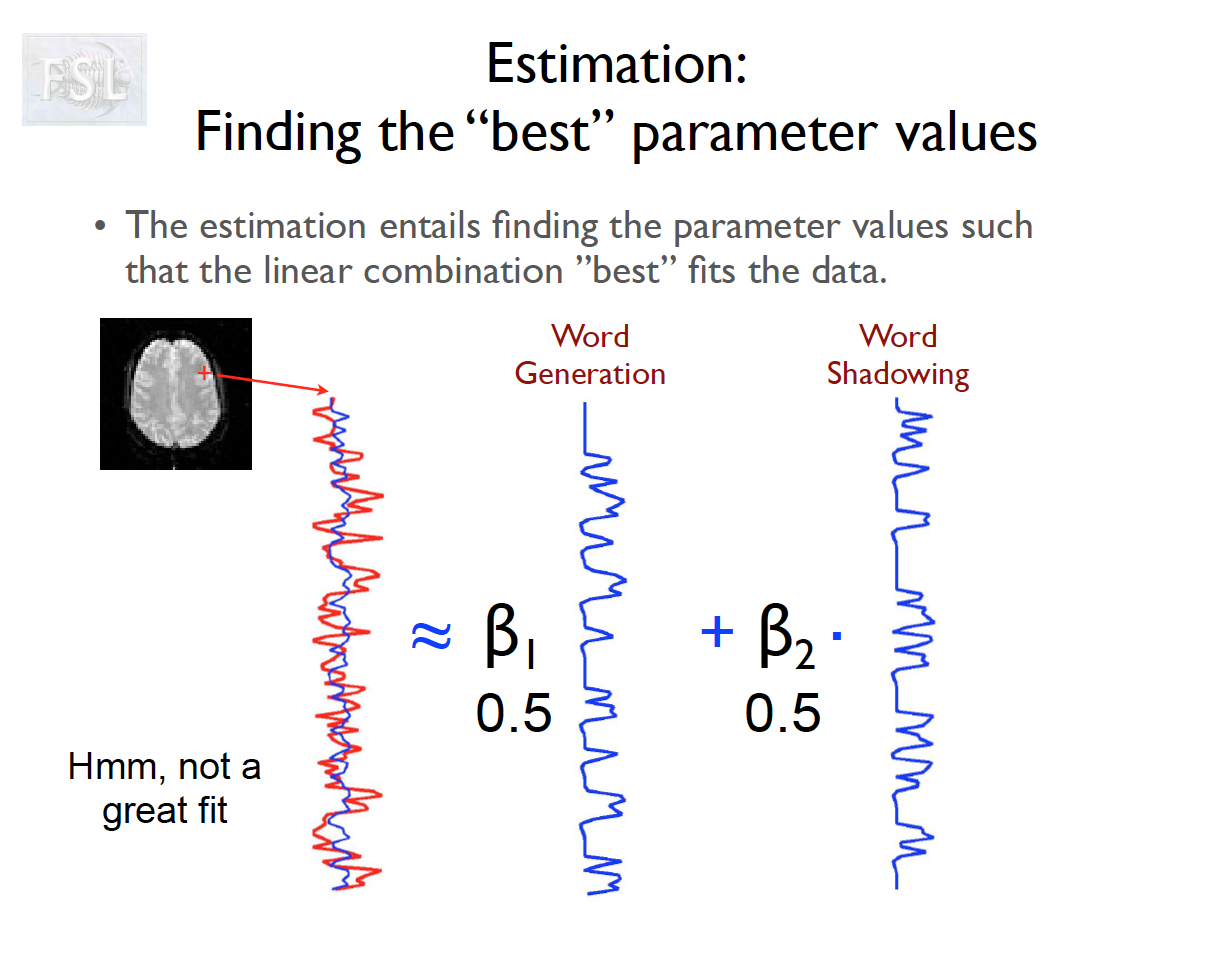
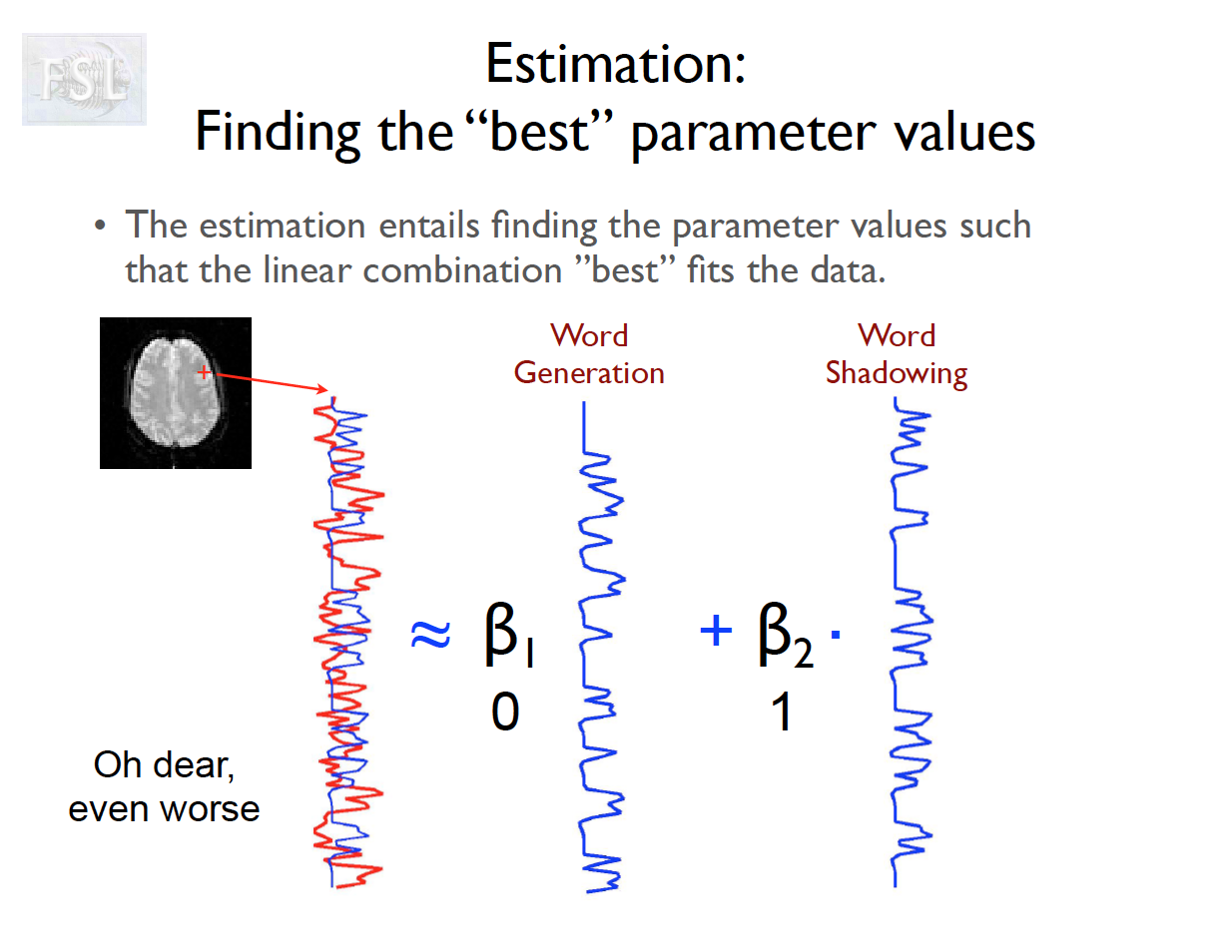
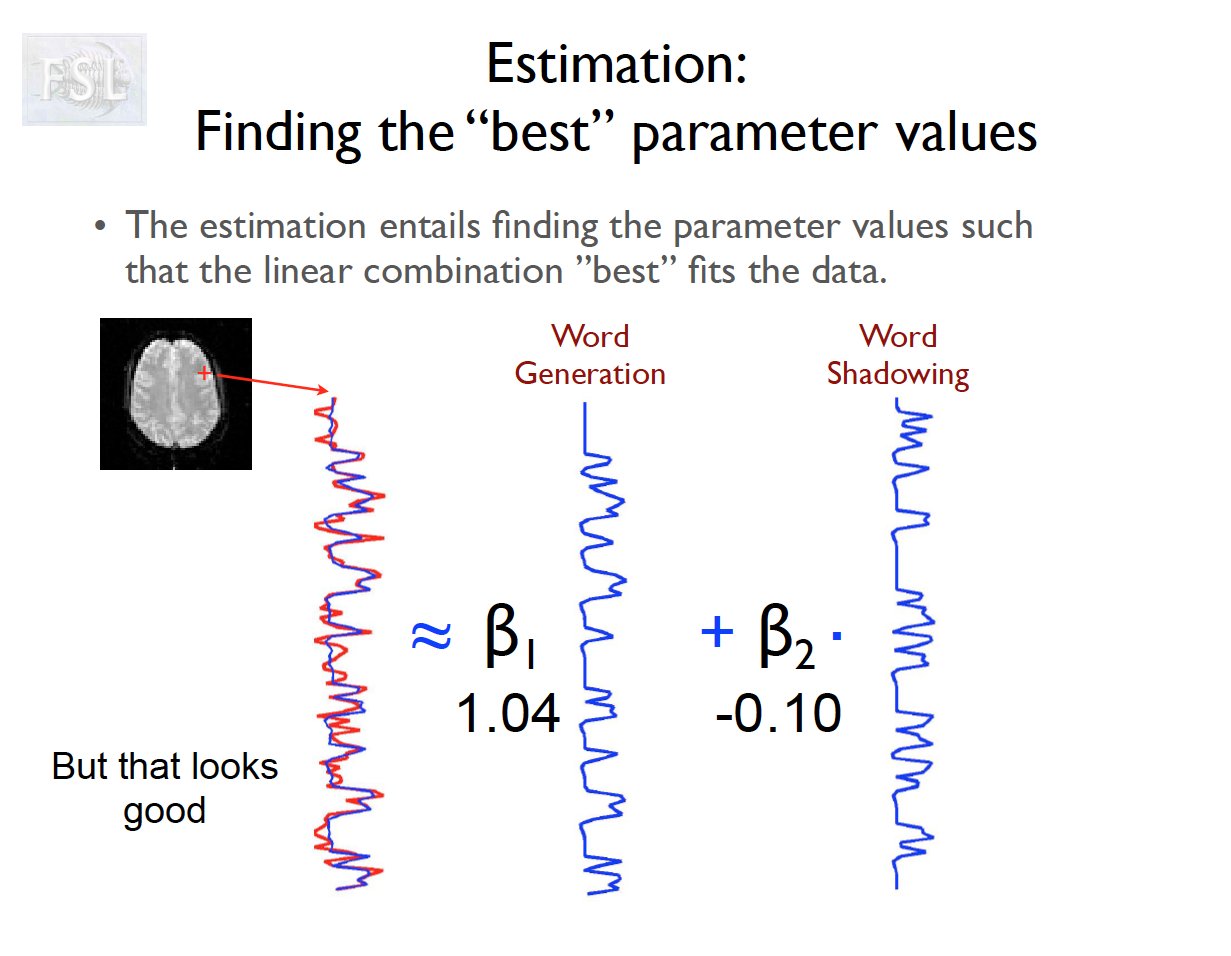
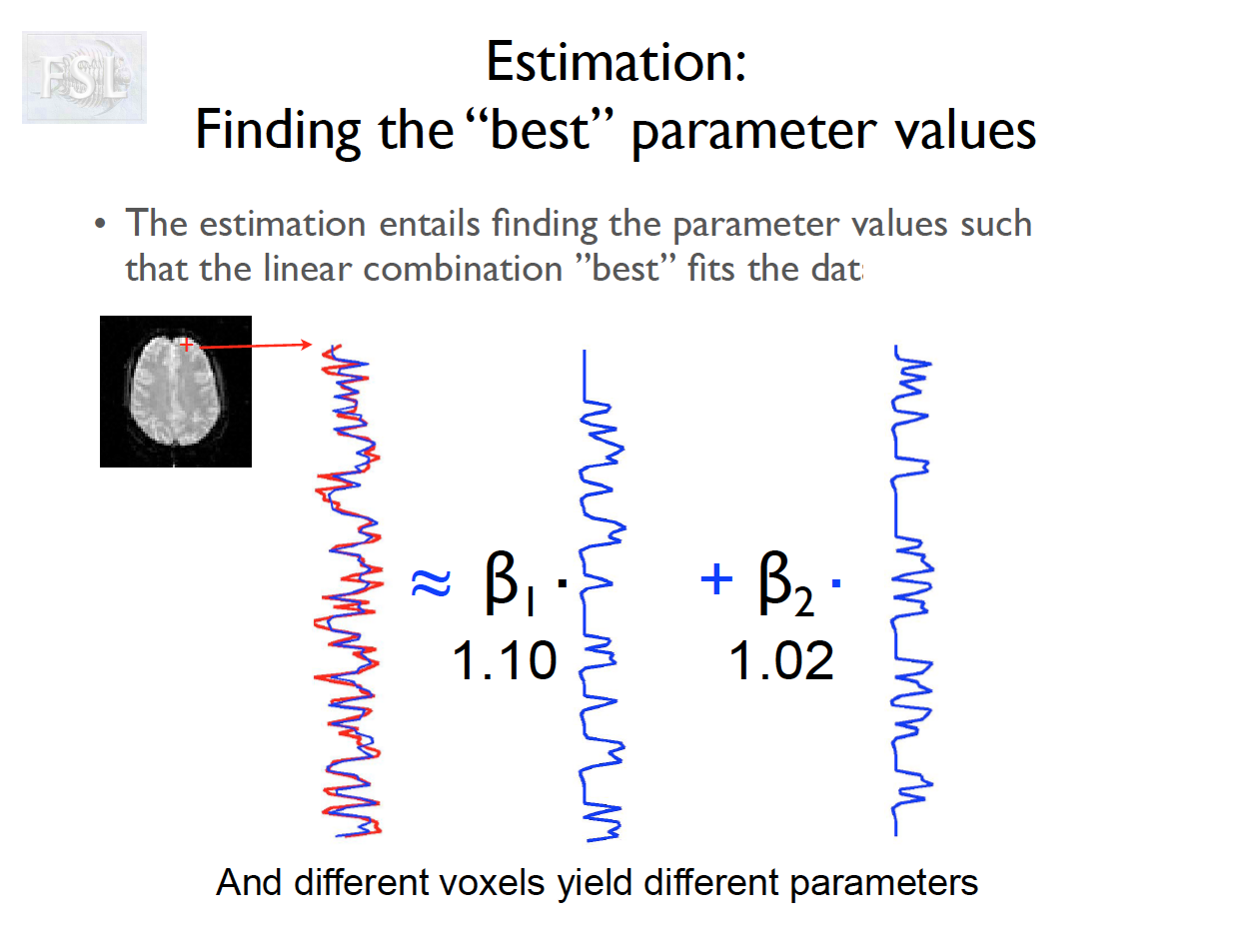
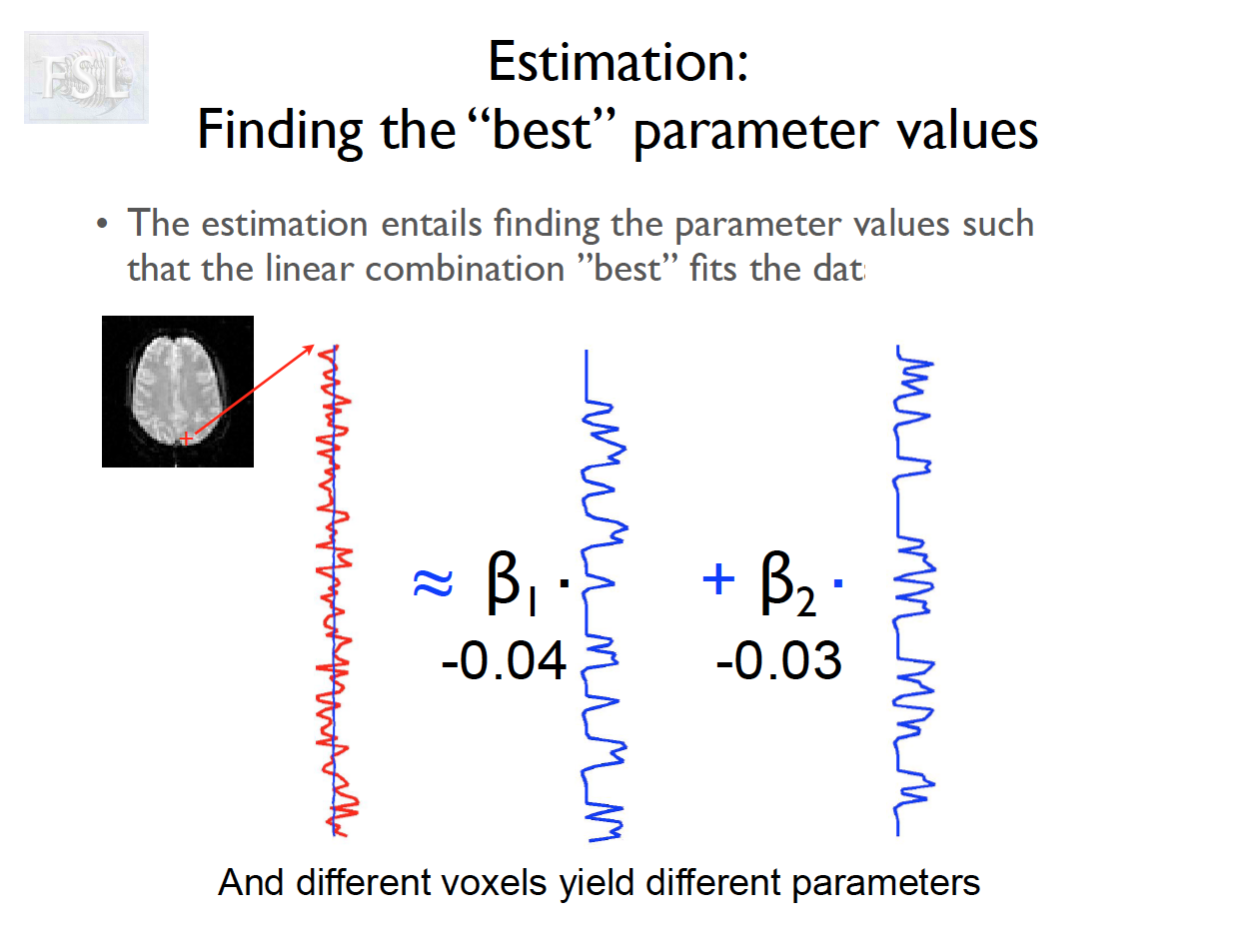
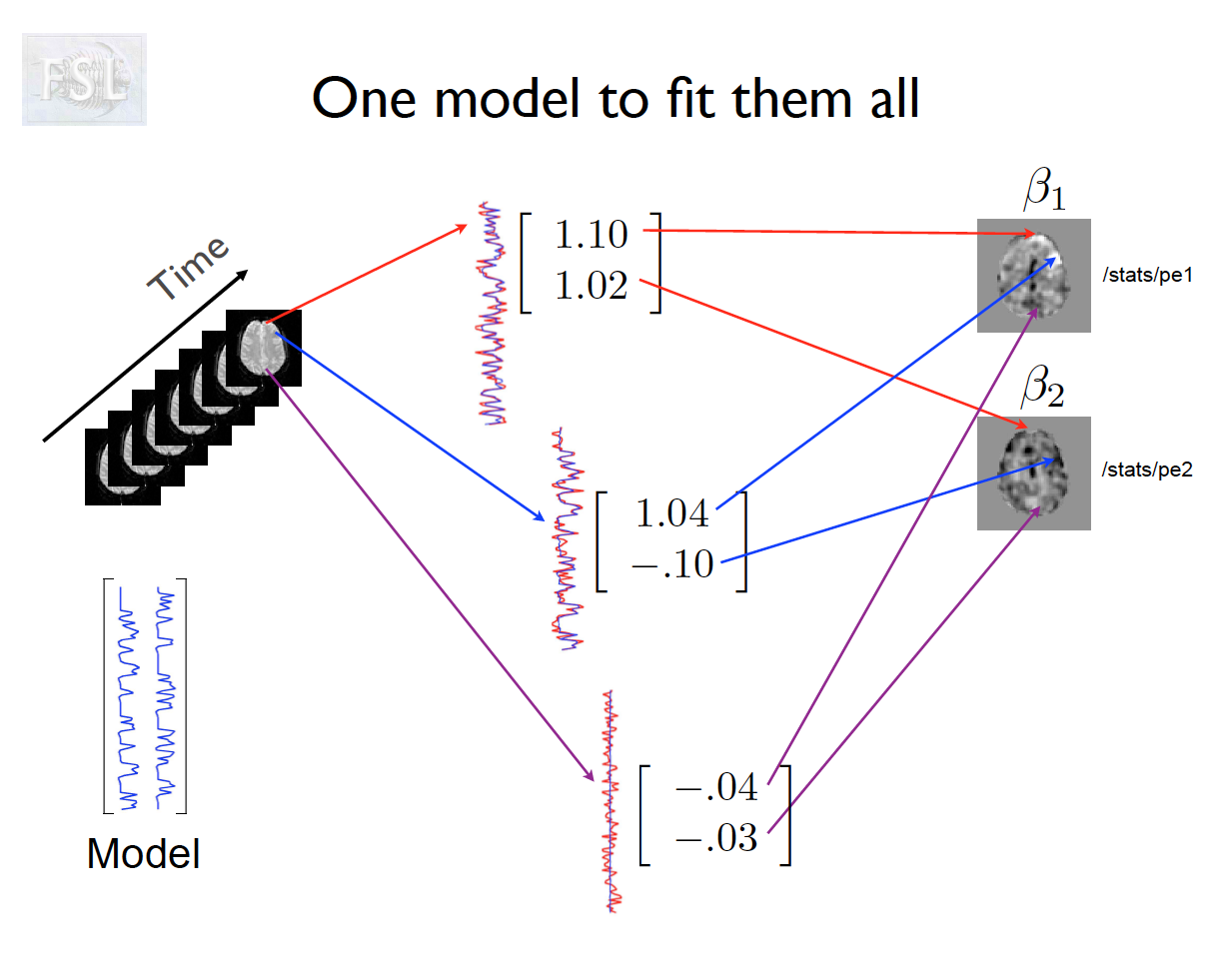
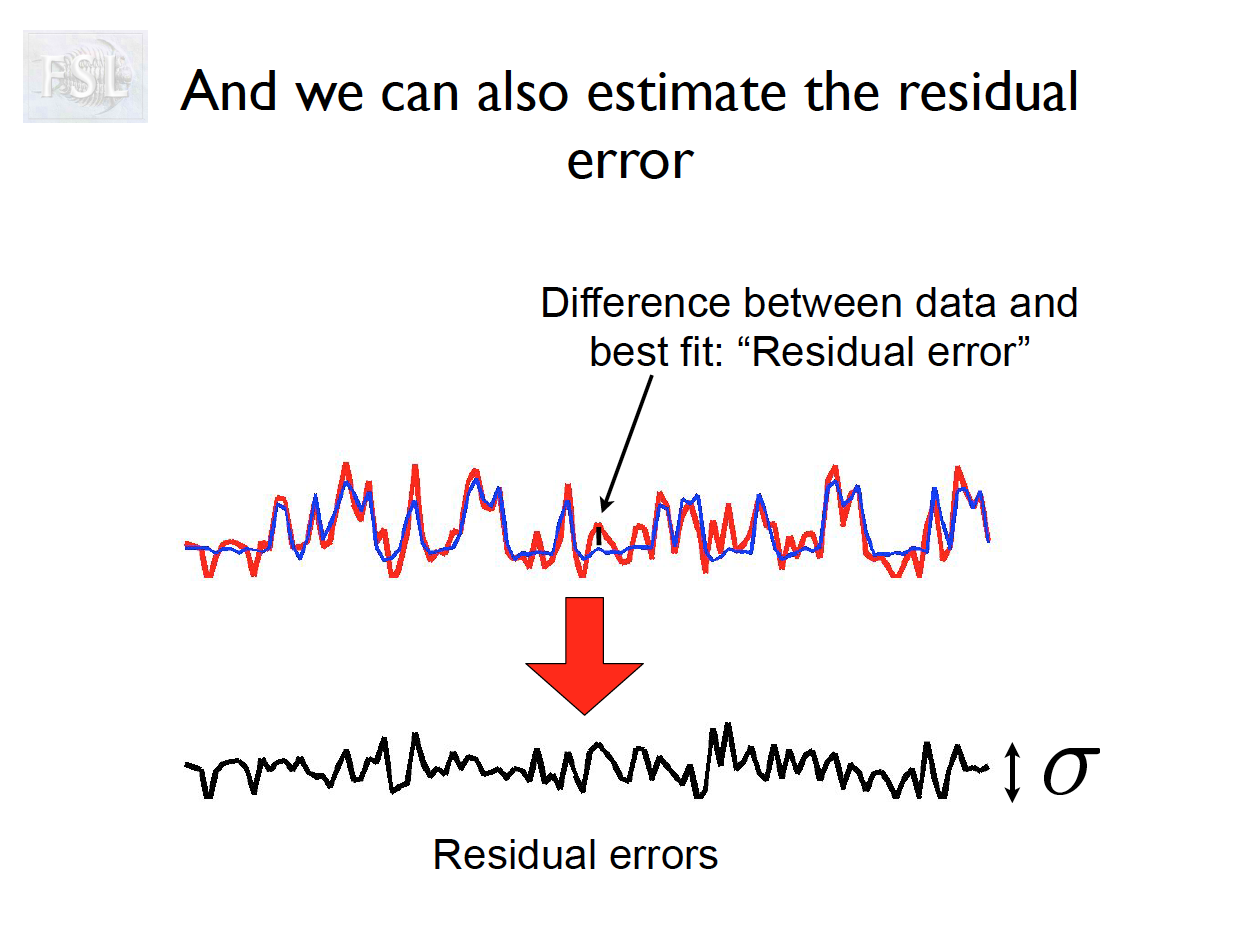
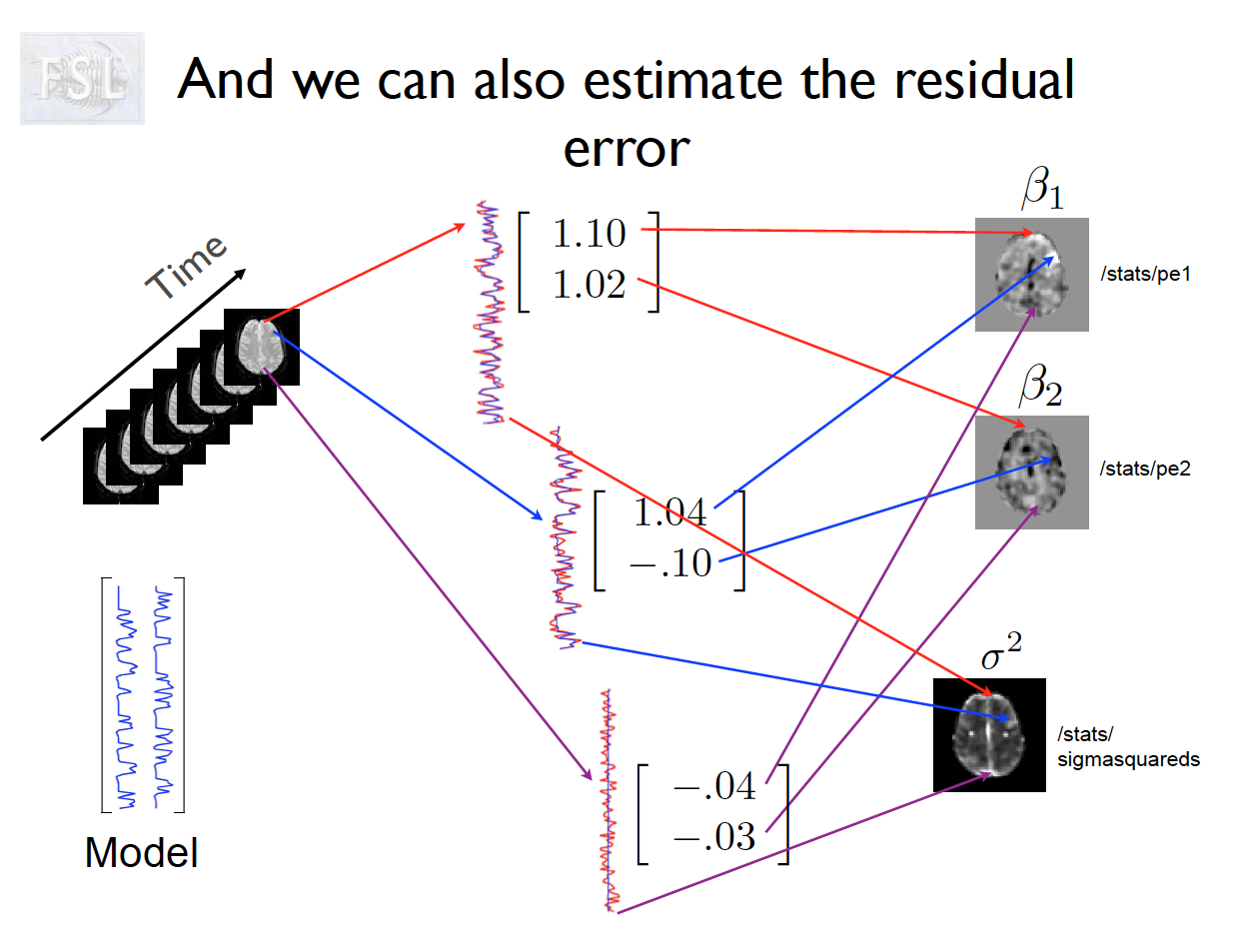
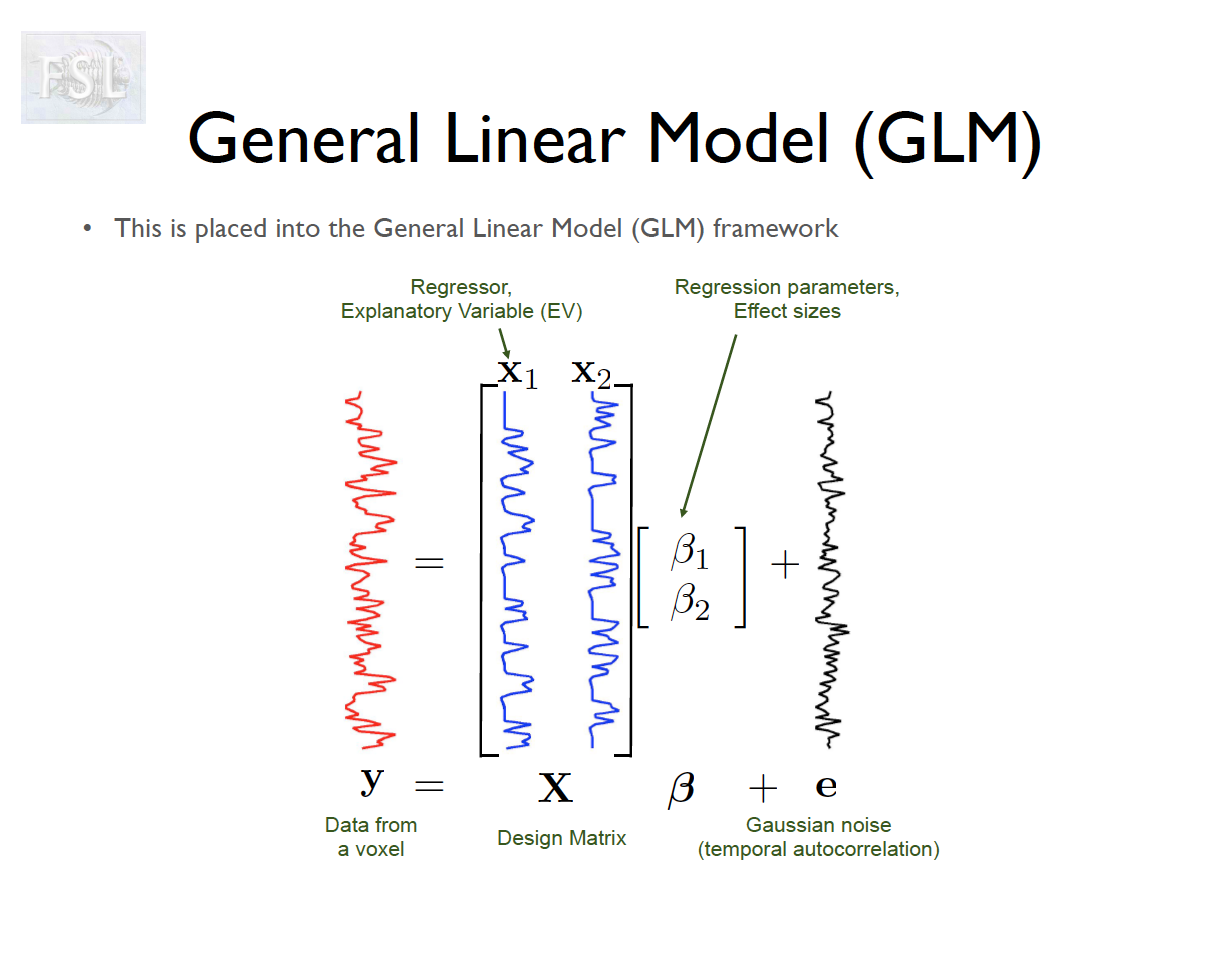
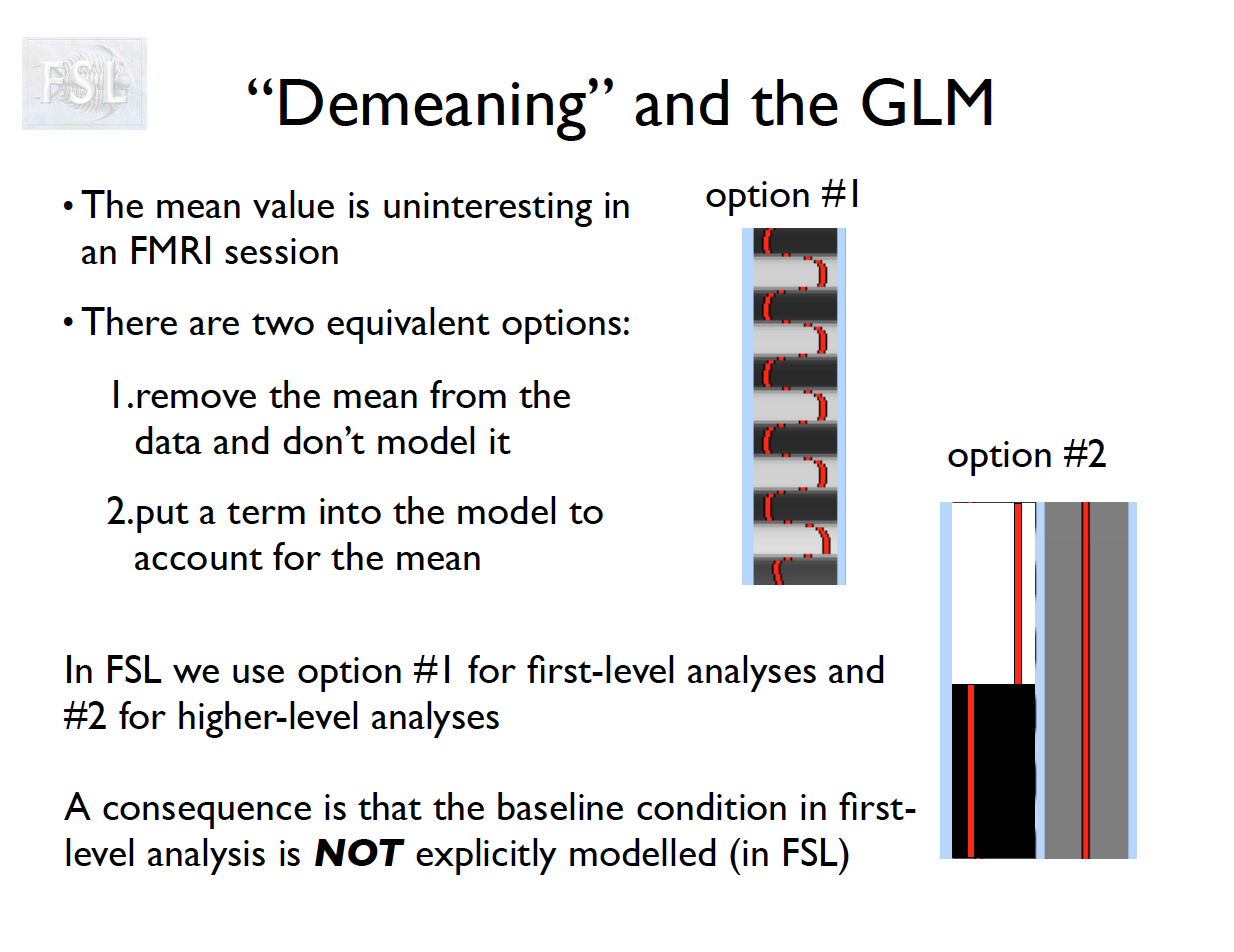
Important points
- task fMRI data independent variables (covariates) are determined by task stimulus series
- And assume hemodynamic response function (double Gamma HRF)
- Models are fit independently at each voxel (spatial location)
- We are only talking about fitting models for one participant, right now.
Time series analysis
Complications with time series
- fMRI time series analysis is more complex than described above
- What critical assumption is violated?
- What will it affect in the model output?
- Think bias and variance of \hat\beta and \hat\sigma_{\hat\beta}^2
- Univariate time series models (such as ARMA) require careful model fit evaluation that is impossible when fitting 150,000 models across the brain
General linear models with prewhitening
- Notes here, based on FILM prewhitening
- For a single voxel v:
y(v) = X\beta(v) + \varepsilon(v)
- Can ignore v, since we are applying across all voxels separately
- X \in \mathbb{R}^{T \times p}: Design matrix (task regressors, confounds)
- \beta \in \mathbb{R}^p: Parameter vector
- \varepsilon \in \mathbb{R}^T: Error
- Assumptions: \mathbb{E}[\varepsilon] = 0, \mathrm{Cov}(\varepsilon ) = \sigma^2V (nonindependent errors)
- If V treated as proportional to the identity matrix, it will lead to inflated false positive rates (Woolrich et al., 2001)
Naive analysis:
What is the expected value of the least squares estimator? \mathbb{E}(\hat \beta) = What is the variance of the least squares estimator? \mathrm{Var}(\hat \beta) =
- A naive variance estimator \hat\sigma^2 (X^TX)^{-1} will be biased.
- Standard errors and test statistics will be biased.
- What is a solution?
Prewhitening Concept
- Solution: transform model so residuals are uncorrelated
Given:
y = X\beta + \varepsilon, \quad \mathrm{Var}(\varepsilon) = \sigma^2V
Let K be a square root matrix of V such that:
K V K^T = I
- Apply K to both sides:
K y = K X \beta + K \varepsilon
- What is the marginal distribution of Ky?
- This is prewhitening: transforms correlated errors into “white noise”
- As you’ve seen, fMRI data are also high-pass filtered to remove low-frequency components.
- The notation in the paper assumes this was already done, resulting in the covariance V
- New Problem: V and K are unknown and need to be estimated.
FILM Approach (Woolrich et al., 2001)
- FMRIB’s Improved Linear Model:
- One-step prewhitening to estimate V
- Many potential time series models to choose from
- FILM models autocorrelation with voxel-wise tapered estimates
Features of time series data
- Auto-correlation
- Frequency domain interpretation
Ways to estimate V
- For an arbitrary time series x(t) we can compute some estimators of the autocorrelation
- Raw estimator for correlation with lag \tau
r_{xx}(\tau) = \frac{1}{\hat\sigma^2} \sum_{t=1}^{N-\tau} x(t) x(t+\tau)/(N-\tau)
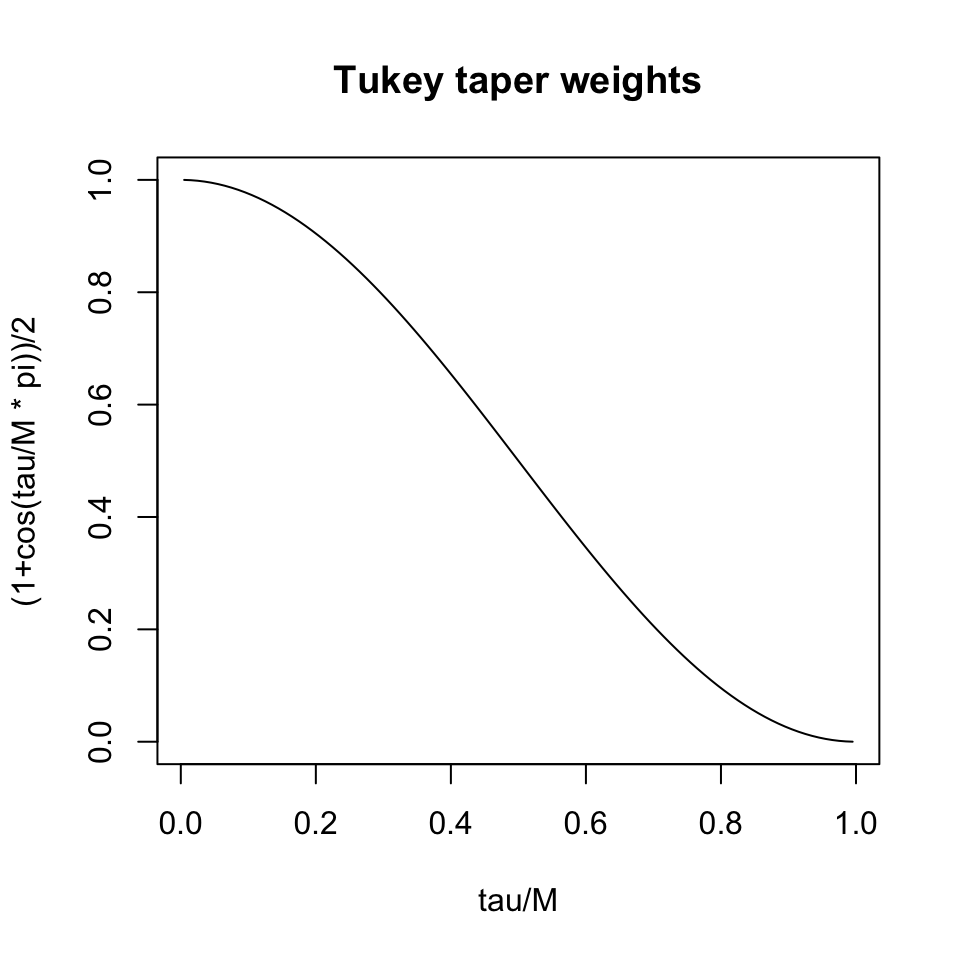
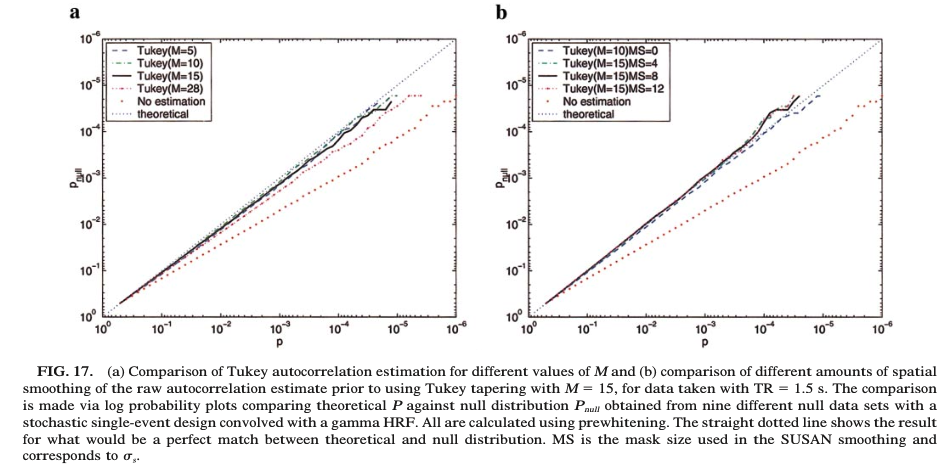
- Tapered estimators, such as the Tukey single-tapering
\hat\rho_{xx}(\tau) = \frac{1}{2}\left( 1 + \cos\left(\frac{\pi \tau}{M}\right) \right) r_{xx}(\tau)
for \tau<M and zero otherwise.
Others not discussed here (see Woolrich et al., 2001)
Matrix V can be filled in with these empirical estimators as a Toeplitz matrix
If \beta is known, the formulas are computed with \epsilon = y - X\beta
Alternatively, we could use \hat\beta, giving the residuals r = (I- X(X^T X)^{-1} X^T) y
What is the autocorrelation of the residuals?
\mathrm{Cov}(r) =
- It is not quite V and V cannot be directly estimated from r. Woolridge et al suggested it is close enough.
Iterative procedure for prewhitening
- \hat\beta is not the BLUE when the covariance of \epsilon is V
- The BLUE is X^T V^{-1} X)^{-1} X^T V^{-1} y (not possible to compute this)
- After obtaining estimate for V (and or K), fit OLS:
\hat{\beta}_1 = (X^T \hat V^{-1} X)^{-1} X^T \hat V^{-1} y
- Then the we could compute a new \hat V and iterate until convergence.
- This would take ages across 150,000 voxels, so instead, they just recompute \hat \beta_1 once
Evaluation
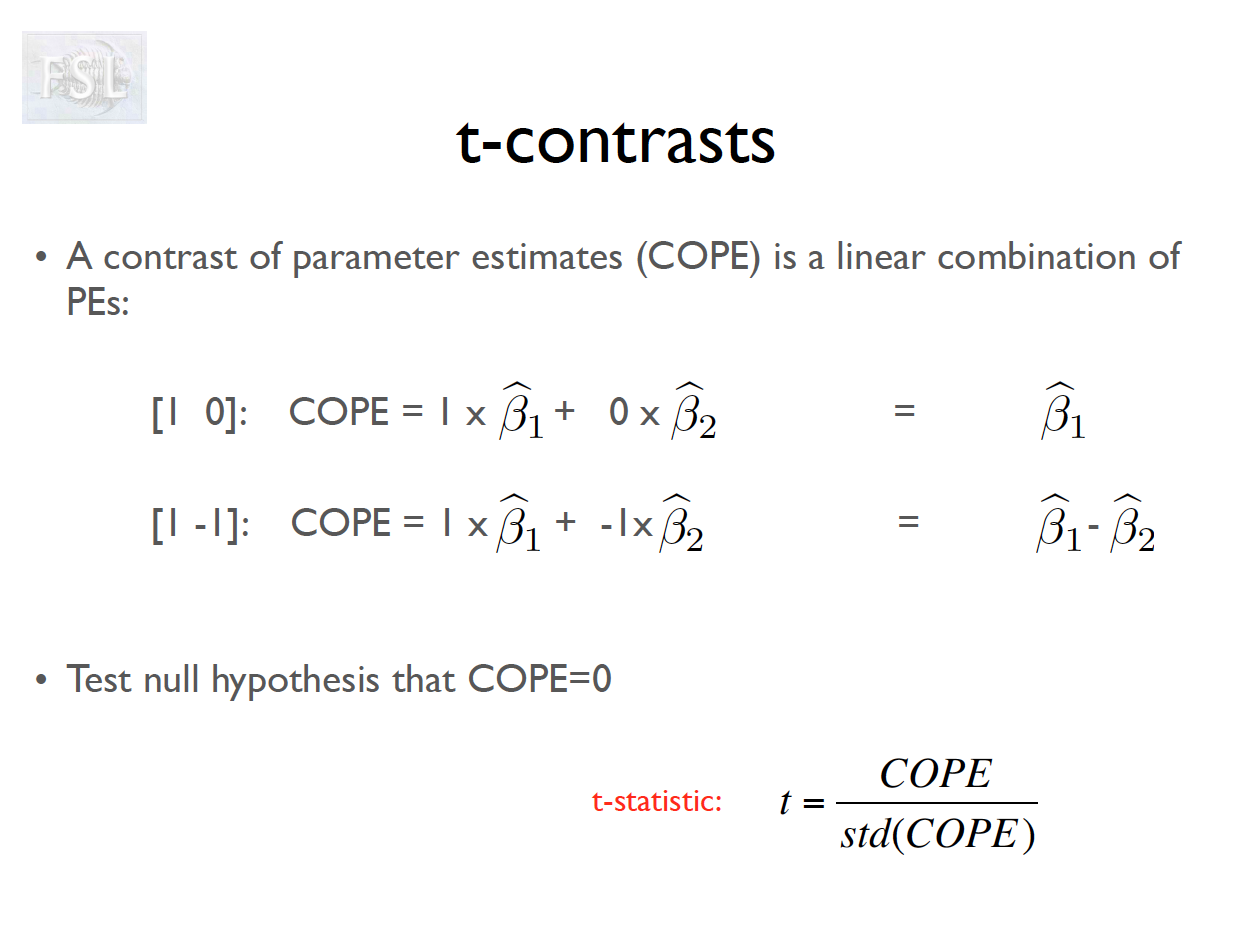
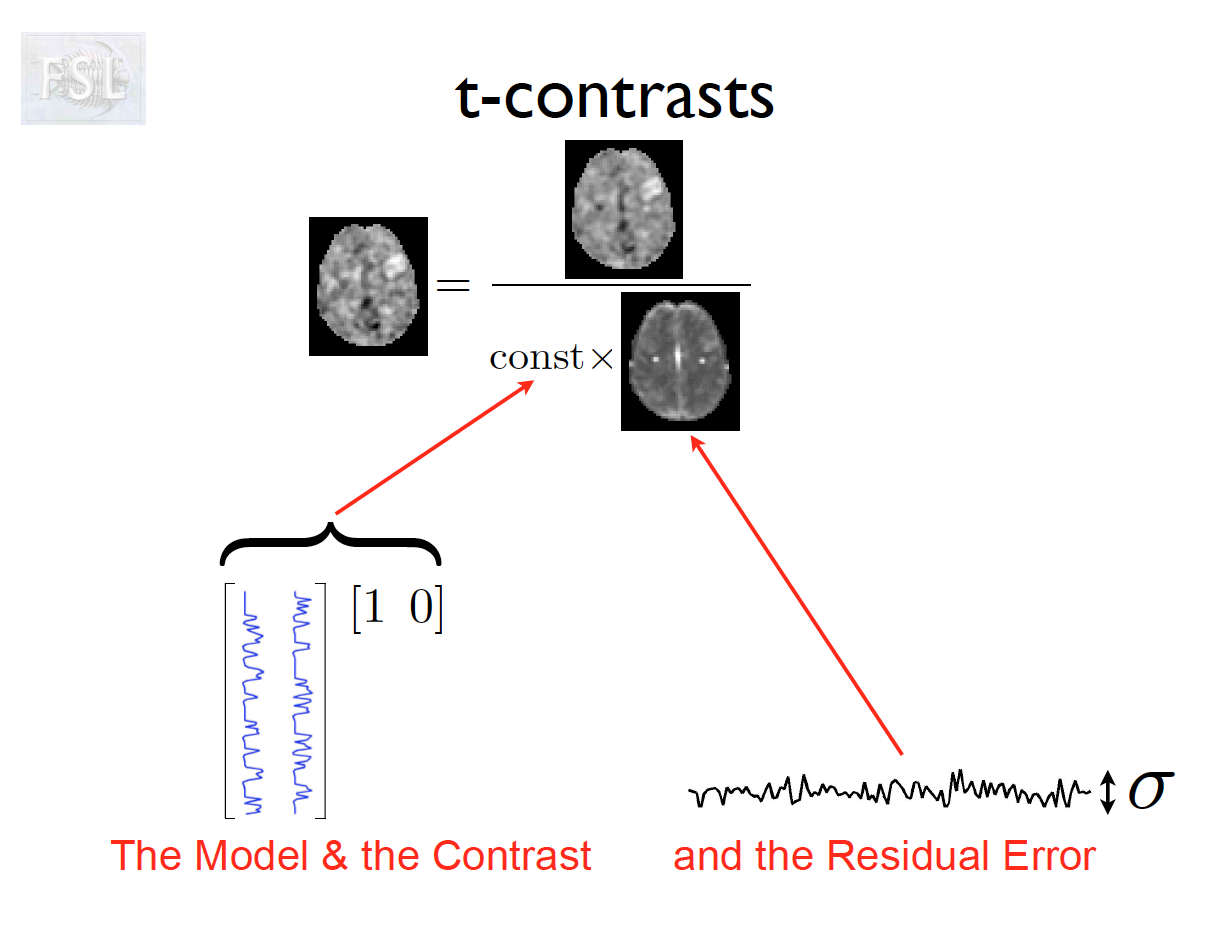
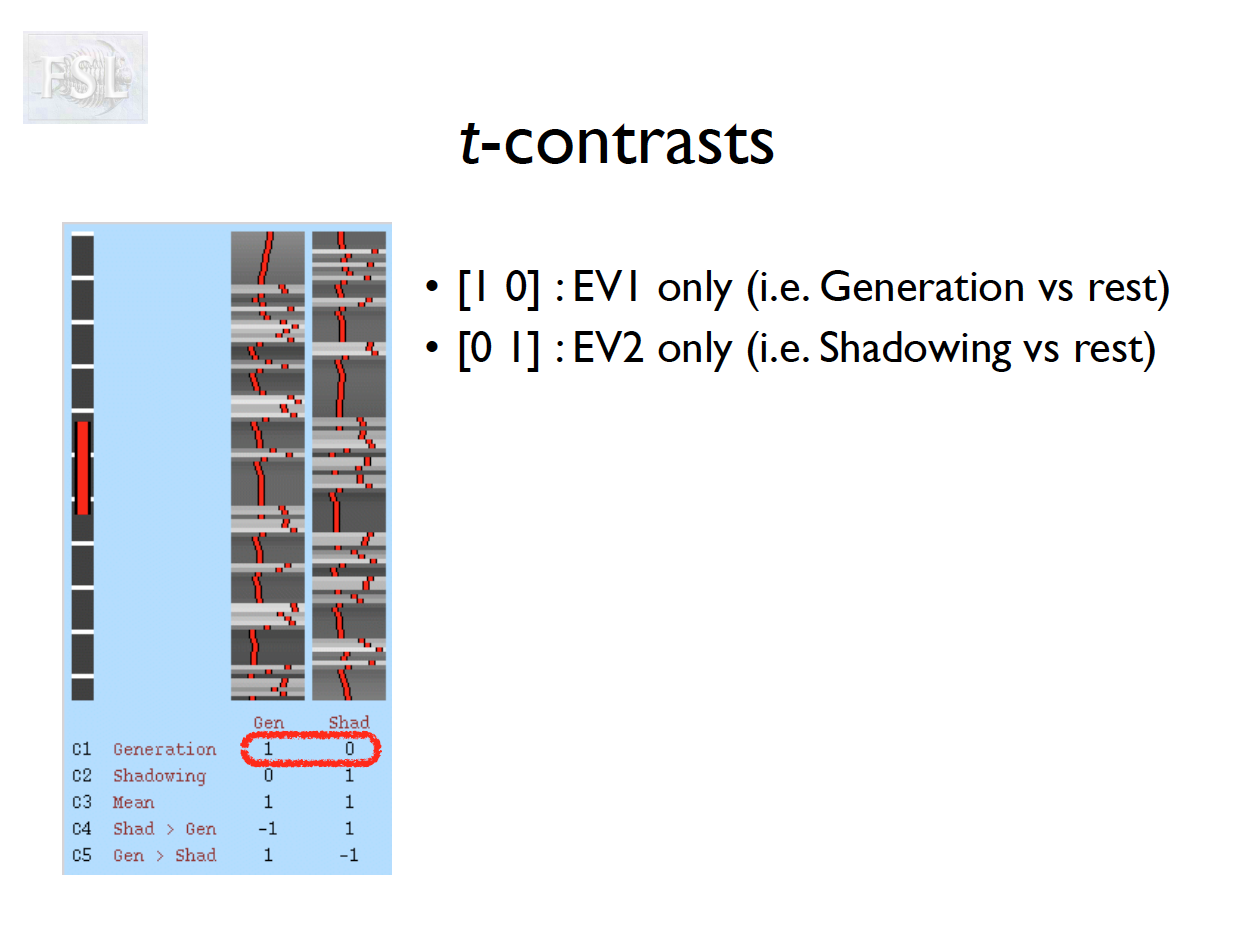
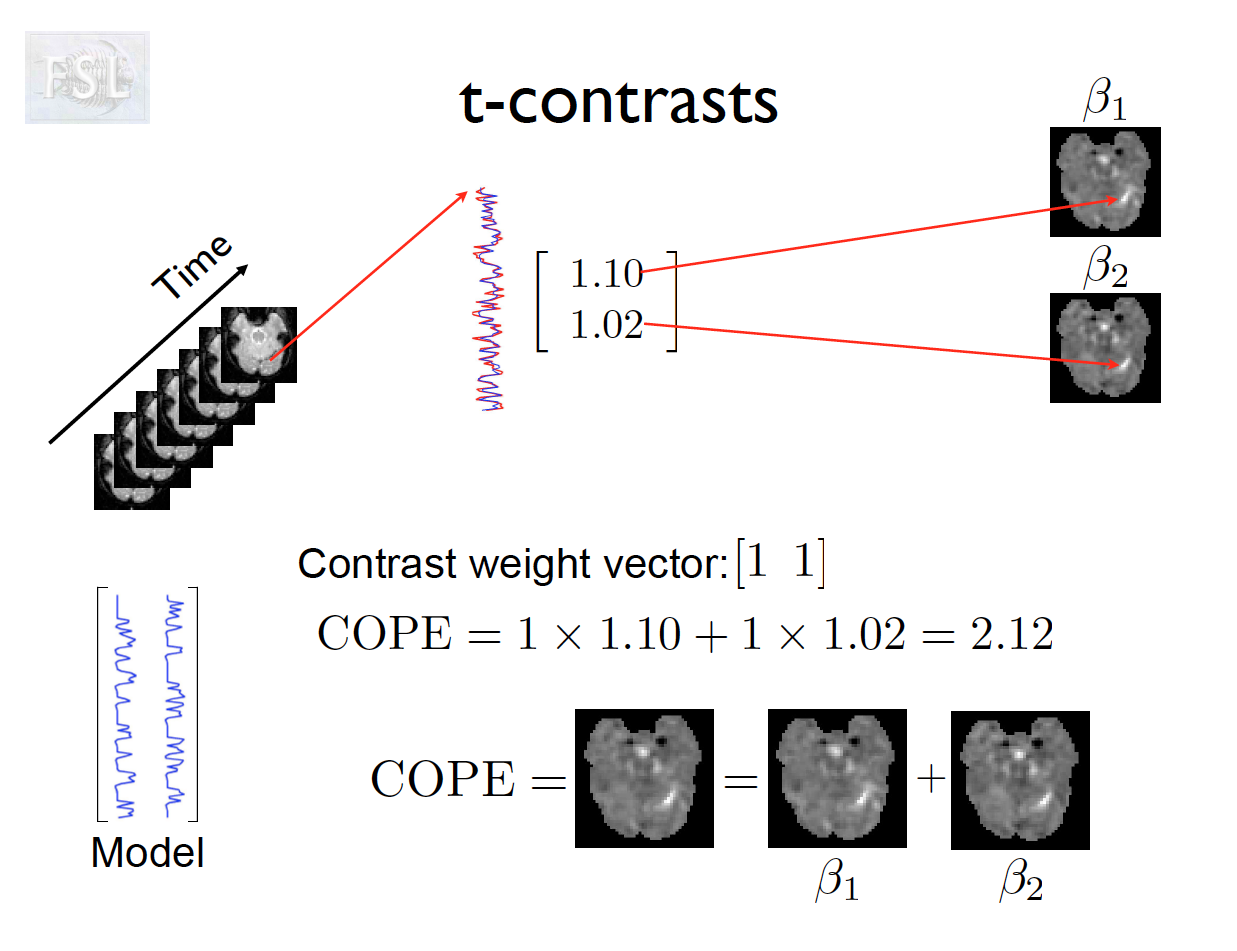
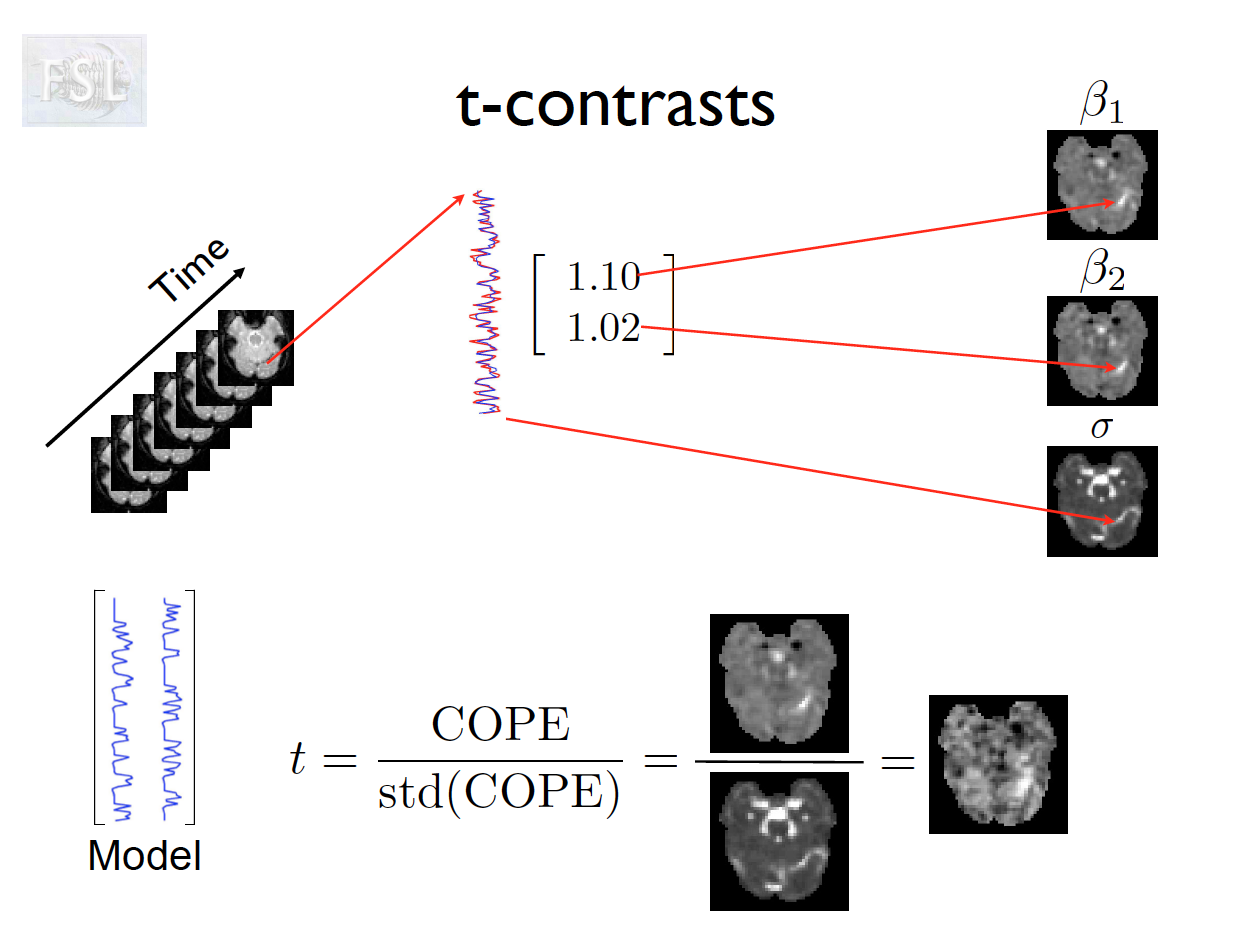
- Contrasts (T- and F-statistics) can be computed:
t = \frac{c^T \hat{\beta}}{\hat{\sigma} \sqrt{c^T (X^T \hat V^{-1} X)^{-1} c}}
and should be approximately normally distributed under the null, in large samples.
References
- Woolrich MW, Ripley BD, Brady M, Smith SM. (2001) Temporal autocorrelation in univariate linear modeling of fMRI data. NeuroImage, 14(6):1370–1386.
- Friston KJ, et al. (1995) Statistical parametric maps in functional imaging: a general linear approach. Human Brain Mapping, 2(4):189–210.
Frequency domain transformations
- e.g. Fourier and Wavelet transformations
- Used a lot in imaging for spatial and temporal frequency domain transformations
- Processing and analysis in the frequency domain has a lot of advantages
- I won’t cover it here, as I’m not an expert, but we could come back to it
Despicable Me
- Interactive: we are going to finish first-level analyses of the Despicable Me dataset
- You will run batch processing on a subset of participants and do QA for the next assignment
Naturalistic viewing task Despicable Me
- We will analyze Naturalistic viewing data
- Naturalistic viewing of Despicable Me (Furtado 2024)
- Despicable Me clip on Netflix 1:02:09 - 1:12:09

Stimulus time series
- Naturalistic viewing task does not have strict “events” that occur during the task
- Some recent research created continuous emotional ratings for the Despicable Me clip
- We can use these to define particular “blocks” of particular emotional states during the clip
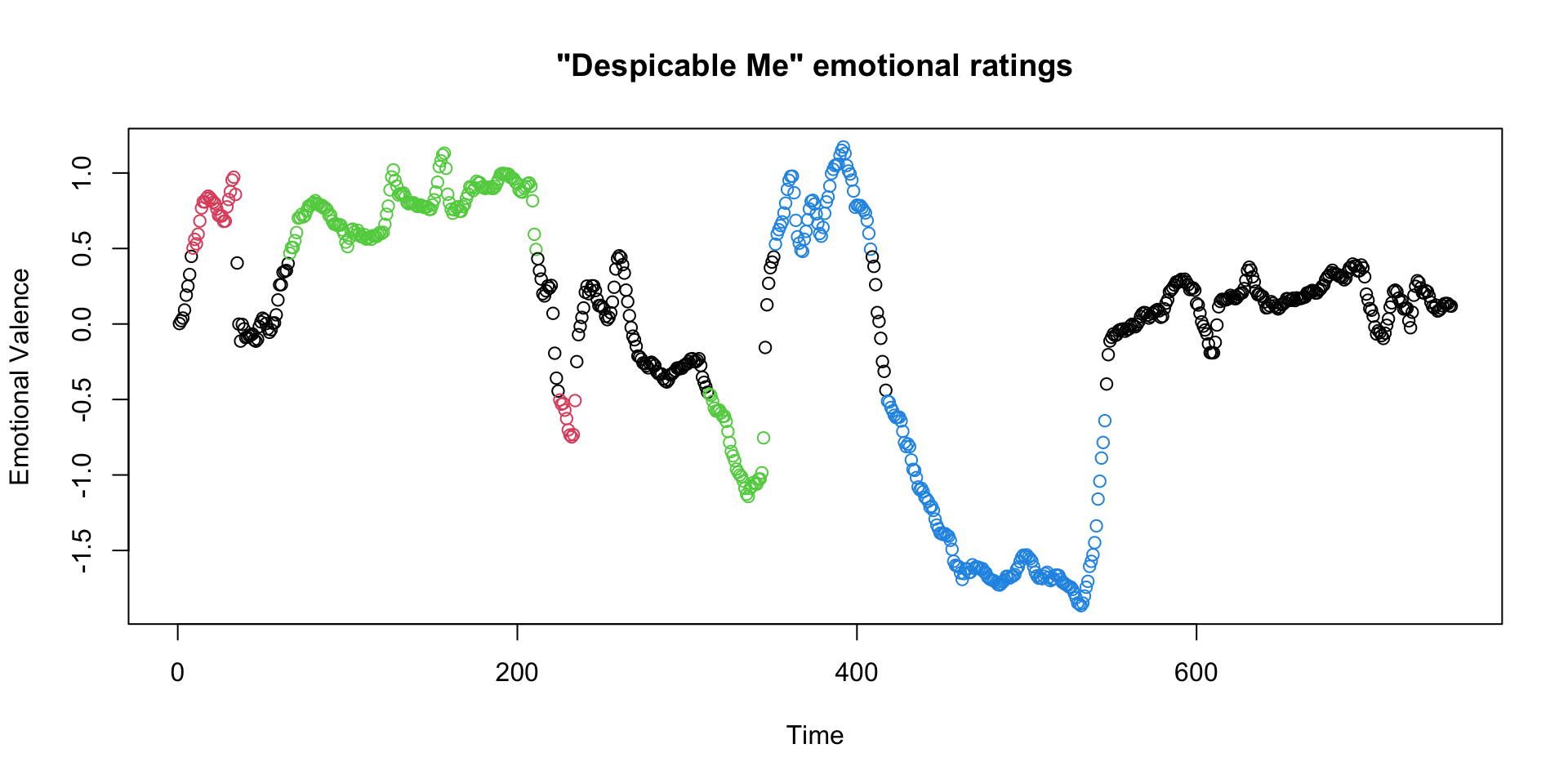
Input into FSL FEAT GUI
- The code below creates a file with the stimulus time series needed for FSL’s FEAT GUI
# path to fsf file for preprocessing for first participant
# /Users/vandeks/Library/CloudStorage/Box-Box/SMN/data/RBC/HBN_BIDS/sub-NDARAA306NT2/ses-HBNsiteRU/func/sub-NDARAA306NT2_ses-HBNsiteRU_task-movieDM_bold.feat/design.fsf
TR = 0.8
pos = rle(dm$positive)
pos$lengths[1] = (pos$lengths[1] - 6)
# first one starts at zero
pos$starts = c(0, cumsum(pos$lengths)[-length(pos$lengths)]+1)
# output needed by FSL
threeCol = data.frame(onset=pos$starts*TR, duration=pos$lengths*TR, value=pos$values)
write.table(threeCol[threeCol$value==1,], row.name=FALSE, sep=' ', col.names = FALSE, file='../data/RBC/stimulusTimeSeries/despicableMe/positiveStimulus.txt')
# /Users/vandeks/Library/CloudStorage/Box-Box/SMN/data//RBC/stimulusTimeSeries/despicableMe/positiveStimulus.txt- To input the stimulus time series into FSL, we provide a 3 column text file with the following info
- Onset, Duration, Value
- Include temporal derivative and apply temporal filtering.
Create a similar stimulus time series for the negative emotional blocks
Design setup

Design matrix
Assigning participants to students
- Sites have different scan parameters, which means we need to process the data differently for each site
- Here, we’ll assign each of you a chunk of participants to process from the same site
- (More on site harmonization later)
resting state fMRI preprocessing
A lot of this preprocessing occurs in a regression framework
Recap of task fMRI processing
- Inhomogeneity correction
- Brain extraction (via structural image)
- Registration
- Motion correction
- Slice timing correction
- Distortion correction (if field map available)
- Temporal filtering & confound regression
- Spatial smoothing
rs-fMRI unique processing
- rs-fMRI is highly sensitive to motion artifact and noise
- More aggressive preprocessing steps:
- Scrubbing – removing/modeling out high-motion time points
- Intensive nuisance covariate removal (24 parameters)
- Band-pass filtering (removing high frequency components as well)
- Independent Components Analysis (ICA) denoising – potential alternative to regression
References Ciric et al., 2017 Parkes et al. 2018
Band-pass filtering
- Similar, but keeping only frequencies in a range
- Performed at the same time as nuisance regression
Nuisance regression (related to motion)
- Motion – 24 parameters total
- Translation and rotation – 6 total parameters
- Derivatives – 6 more
- Squares of translation and rotations
- Squares of derivatives
Nuisance regression (nuisance signal)
aCompCor – Anatomical Component Correlation
- In addition to motion covariates
- CSF and white matter are not “active” – regress out signal associated with these
- PCA of the signal in these regions, e.g. Y_{\mathrm{CSF}} \in \mathbb{R}^{T \times V_{\mathrm{CSF} }} \\ Y_{\mathrm{CSF}} = U D W^T Choose the first K components and regress them out of the data
Global signal regression
- Controversial because there is evidence it increases motion-related correlations between distant brain regions
- I am not too familiar why, could be a good advanced topic
- REFs.
Scrubbing
“Dummy”/“One hot” coding of scans where participant move a lot
In the end, the preprocessing regression design matrix for rs-fMRI might look something like
\begin{bmatrix} \vert & \vert & \vert& \vert \\ \text{Temp filt} & \text{Motion} & \text{aCompCor} & \text{Dummy variables} \\ \vert & \vert & \vert & \vert \end{bmatrix}
rs-fMRI before and after preprocessing
- Histogram of signal changes
- Positive/negative correlations are relative to preprocessing
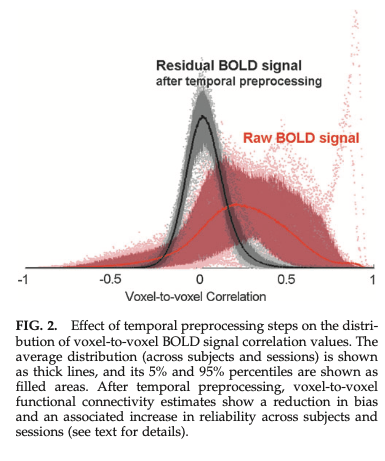
Whitfield-Gabrieli 2012
rs-fMRI first-level analysis
- Pretty straightforward after preprocessing
- Functional connectivity is defined as Pearson correlation between two locations (R(v,t) is the residuals from the preprocessing)
\hat \rho(v,w) = \frac{\sum_{t=1}^T R(v, t) R(w, t)}{\sqrt{\sum_{t=1}^T R(v, t) ^2 \times \sum_{t=1}^T R(w, t)^2 } } * Correlations are Fisher Z-transformed so that they are approximately normal
Z(v,w) = \mathrm{atanh}(\hat \rho(v,w))
- Types of analyses
- Network – all voxel-to-voxel or region-to-region correlations
- Seed-based – Correlation of preselected seed region with entire image (image-valued)
Overview of rs-fMRI
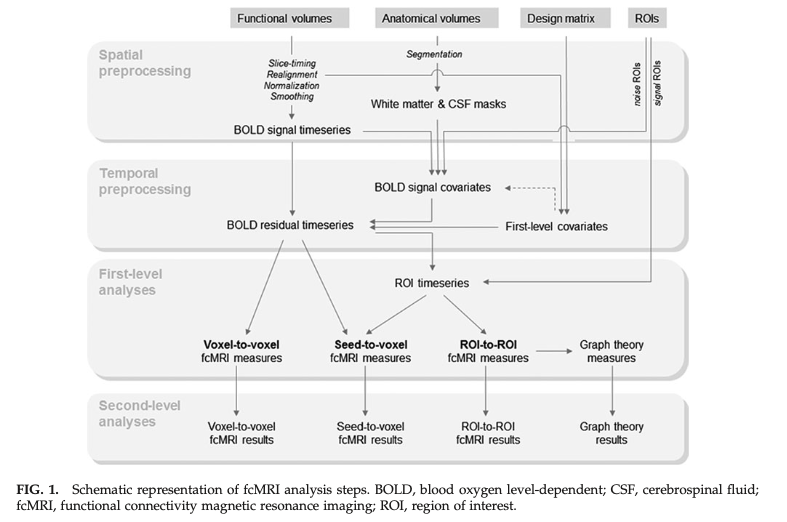
Whitfield-Gabrieli 2012